Code testing#

Looking back at our script from the section before, ie the one you just brought up to code concerning comments and formatting based on guidelines, there is at least one other big question we have to address…
How do we know that the code does what it is supposed to be doing?
Now you might think “What do you mean? It is running the things I wrote down, done.”. However, the reality looks different…
Background#
Generally, there are two major reasons why it is of the utmost importance to check and evaluate code:
Mistakes made while coding
Code instability and changes
Let’s have a quick look at both.
Mistakes made while coding#
It is very, very easy to make mistakes when coding. A single misplaced character can cause a program/script’s output to be entirely wrong or vary tremendously from what its expected behavior. This can happen because a plus sign which should have been a minus or one piece of code working in one unit while a piece of code written by another researcher worked in a differnt unit. Everyone makes mistakes, but the results can be catastrophic. Careers can be damaged/ended, vast sums of research funds can be wasted, and valuable time may be lost to exploring incorrect avenues.

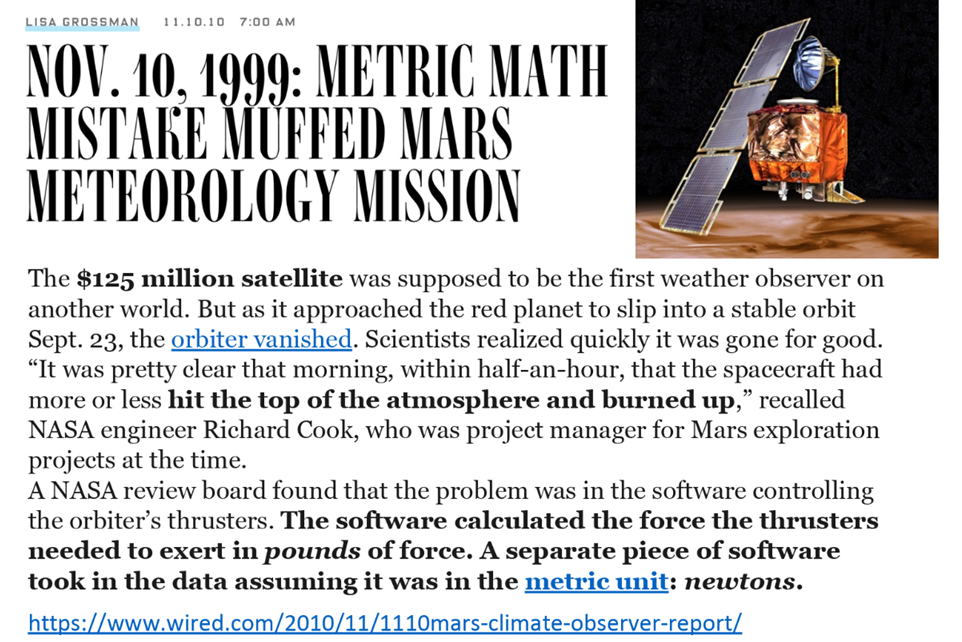


Code instabilities and changes#
The second reason is a also challening but in a different way…The code you are using and writing is affected by underlying numerical instabilities and changes during development. Just have a look at this example.
print(sum([0.001 for _ in range(1000)]))
1.0000000000000007
Regarding the first, there are intrinsic numerical erros & instabilities that may lead unstable functions towards distinct local minima. This phenomenon is only aggravated by prominent differences between OS.
Concerning the second, a lot of the code you are using is going to be part of packages and modules that are developed and maintained by other people. Along this process, the code, e.g. a function, you are using is going to change, more or less prominently. It could be a rounding change or complete change of inputs and outputs. Either or, the effects on your application and/or pipeline might be significant and most importantely: unbeknownst to you.
This is why software and code tests are vital! Ie, to ensure the expected outcome and check, as well as evaluate changes along the development process.
Motivation#
save time and effort#
Even if problems in a program are caught before research is published it can be difficult to figure out what results are contaminated and must be re-done. This represents a huge loss of time and effort. Catching these problems as early as possible minimises the amount of work it takes to fix them, and for most researchers time is by far their most scarce resource. You should not skip writing tests because you are short on time, you should write tests because you are short on time. Researchers cannot afford to have months or years of work go down the drain, and they can’t afford to repeatedly manually check every little detail of a program that might be hundreds or hundreds of thousands of lines long. Writing tests to do it for you is the time-saving option, and it’s the safe option.

long-term confidence#
As researchers write code they generally do some tests as they go along, often by adding in print statements and checking the output. However, these tests are often thrown away as soon as they pass and are no longer present to check what they were intended to check. It is comparatively very little work to place these tests in functions and keep them so they can be run at any time in the future. The additional labour is minimal, the time saved and safeguards provided are invaluable. Further, by formalizing the testing process into a suite of tests that can be run independently and automatically, you provide a much greater degree of confidence that the software behaves correctly and increase the likelihood that defects will be found.

peace of mind#
Testing also affords researchers much more peace of mind when working on/improving a project. After changing their code a researcher will want to check that their changes or fixes have not broken anything. Providing researchers with a fail-fast environment allows the rapid identification of failures introduced by changes to the code. The alternative, of the researcher writing and running whatever small tests they have time for is far inferior to a good testing suite which can thoroughly check the code.

better code from the start#
Another benefit of writing tests is that it typically forces a researcher to write cleaner, more modular code as such code is far easier to write tests for, leading to an improvement in code quality. Good quality code is far easier (and altogether more pleasant) to work with than tangled rat’s nests of code I’m sure we’ve all come across (and, let’s be honest, written). This point is expanded upon in the section Unit Testing.

Research software#
As well as advantaging individual researchers testing also benefits research as a whole. It makes research more reproducible by answering the question “how do we even know this code works”. If tests are never saved, just done and deleted the proof cannot be reproduced easily.
Testing also helps prevent valuable grant money being spent on projects that may be partly or wholly flawed due to mistakes in the code. Worse, if mistakes are not at found and the work is published, any subsequent work that builds upon the project will be similarly flawed.
Perhaps the cleanest expression of why testing is important for research as a whole can be found in the Software Sustainability Institute slogan: better software, better research.


Write Tests - Any Tests!#
Starting the process of writing tests can be overwhelming, especially if you have a large code base. Further to that, as mentioned, there are many kinds of tests, and implementing all of them can seem like an impossible mountain to climb. That is why the single most important piece of guidance in this chapter is as follows: write some tests. Testing one tiny thing in a code that’s thousands of lines long is infinitely better than testing nothing in a code that’s thousands of lines long. You may not be able to do everything, but doing something is valuable.
Make improvements where you can, and do your best to include tests with new code you write even if it’s not feasible to write tests for all the code that’s already written.

Run the tests#
The second most important piece of advice in this chapter: run the tests. Having a beautiful, perfect test suite is no use if you rarely run it. Leaving long gaps between test runs makes it more difficult to track down what has gone wrong when a test fails because, a lot of the code will have changed. Also, if it has been weeks or months since tests have been run and they fail, it is difficult or impossible to know which results that have been obtained in the mean time are still valid, and which have to be thrown away as they could have been impacted by the bug.

It is best to automate your testing as far as possible. If each test needs to be run individually then that boring painstaking process is likely to get neglected. This can be done by making use of a testing framework (discussed later). Ideally set your tests up to run at regular intervals, possibly every night.
Consider setting up continuous integration (discussed in the continuous integration sesssion) on your project. This will automatically run your tests each time you make a change to your code and, depending on the continuous integration software you use, will notify you if any of the tests fail.
Consider how long it takes your tests to run#
Some tests, like Unit Testing only test a small piece of code and so typically are very fast. However other kinds of tests, such as System Testing which test the entire code from end to end, may take a long time to run depending on the code. As such it can be obstructive to run the entire test suite after each little bit of work.

In that case it is better to run lighter weight tests such as unit tests frequently, and longer tests only once per day, overnight. It is also good to scale the number of each kind of tests you have in relation to how long they take to run. You should have a lot of unit tests (or other types of tests that are fast) but much fewer tests which take a long time to run.
Document the tests and how to run them#
It is important to provide documentation that describes how to run the tests, both for yourself in case you come back to a project in the future, and for anyone else that may wish to build upon or reproduce your work.

This documentation should also cover subjects such as:
any resources, such as
test datasetfiles that are requiredany
configuration/settingsadjustments needed to run the testswhat
software(such astesting frameworks) need to be installed
Ideally, you would provide scripts to set up and configure any resources that are needed.
Test Realistic Cases#
Make the cases you test as realistic as possible. If for example, you have dummy data to run tests on you should make sure that data is as similar as possible to the actual data. If your actual data is messy with a lot of null values, so should your test dataset be.
Use a Testing Framework#
There are tools available to make writing and running tests easier, these are known as testing frameworks. Find one you like, learn about the features it offers, and make use of them. A very common testing framework for python is pytest.
Coverage#
Code coverage is a measure of how much of your code is “covered” by tests. More precisely it a measure of how much of your code is run when tests are conducted. So for example, if you have an if statement but only test things where that if statement evaluates to “False” then none of the code in the if block will be run. As a result your code coverage would be < 100%. Code coverage doesn’t include documentation like comments, so adding more documentation doesn’t affect your percentages.
Code coverage gauges how much code your tests run, aiming for as close to 100% as possible without counting documentation. High coverage is ideal, though any testing is beneficial. Various tools and bots measure this across programming languages, e.g. pytest for python. Beware the illusion of good coverage; thorough testing involves multiple scenarios for the same code, emphasizing testing smaller code chunks for precise logic validation. Testing the same code multiple ways is encouraged for comprehensive assessment.

Use test doubles/stubs/mocking where appropriate#
Use test doubles like stubs or mocks for isolating code in tests. Ensure tests make it easy to pinpoint failures, which can be hard when code depends on external factors like internet connections or objects. For example, a web interaction test might fail due to internet issues, not code bugs. Similarly, a test involving an object might fail because of the object itself, which should have its own tests. Eliminate these dependencies with test doubles, which come in several types:
Dummy objectsareplaceholdersthat aren’t actually used intestingbeyond filling method parameters.# Dummy value to fill in as a placeholder dummy_logger = None def process_data(data, logger): # The function doesn't actually use the logger in this case return f"Processed {data}" print(process_data("input_data", dummy_logger)) # Output: Processed input_data
Fake objectshave simplified, functional implementations, like anin-memory databaseinstead of a real one.# Fake buffer fake_buffer = [] def add_reading(buffer, reading): buffer.append(reading) def get_latest_reading(buffer): return buffer[-1] if buffer else None # Using the fake buffer in a test add_reading(fake_buffer, 25.3) print(get_latest_reading(fake_buffer)) # Output: 25.3
Stubsprovide partial implementations to respond only to specifictestcases and might record call information.# Stub that only provides results for specific test inputs stub_calls = [] # List to record calls def calculate_distance(speed, time): stub_calls.append((speed, time)) # Record the call if speed == 10 and time == 2: return 20 # A specific expected response return None # Using the stub in a test print(calculate_distance(10, 2)) # Output: 20 print(stub_calls) # Output: [(10, 2)]
Mockssimulateinterfacesorclasses, withpredefined outputsformethodcalls, often recording interactions fortestvalidation.from unittest.mock import Mock # Mock function to simulate a complex calculation mock_calculate = Mock(return_value=42) # Using the mock in a test result = mock_calculate(5, 7) print(result) # Output: 42 # Verify the mock was called with specific arguments mock_calculate.assert_called_with(5, 7)
Test doubles replace real dependencies, making tests more focused and reliable. Mocks can be hand-coded or generated with mock frameworks, which allow dynamic behavior definition. A common mock example is a data provider, where a mock simulates the data source to ensure consistent test conditions, contrasting with the real data source used in production.
Code Testing - the basics#
There are a number of different kinds of tests, which will be briefly discussed in the follwing.
Firstly, there are positive tests and negative tests. Positive tests check that something works, for example, testing that a function that multiplies some numbers together outputs the correct answer. Negative tests check that something generates an error when it should. For example, nothing can go quicker than the speed of light, so a plasma physics simulation code may contain a test that an error is outputted if there are any particles faster than this, as it indicates there is a deeper problem in the code.
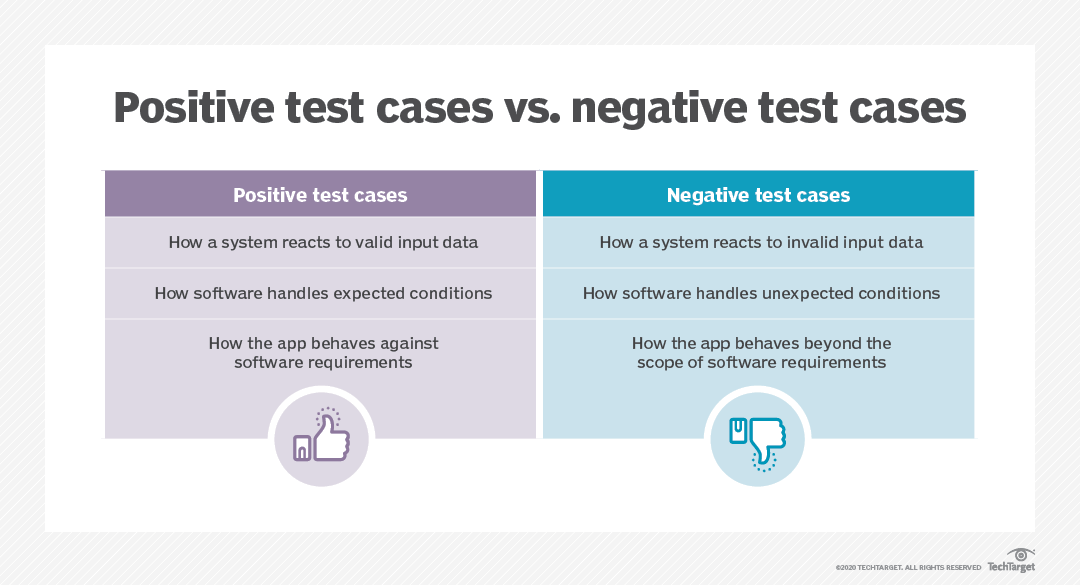
Image credit: TechTarget.
In addition to these two kinds of tests, there are also different levels of tests which test different aspects of a project. These levels are outlined below and both positive and negative tests can be present at any of these levels. A thorough test suite will contain tests at all of these levels (though some levels will need very few).
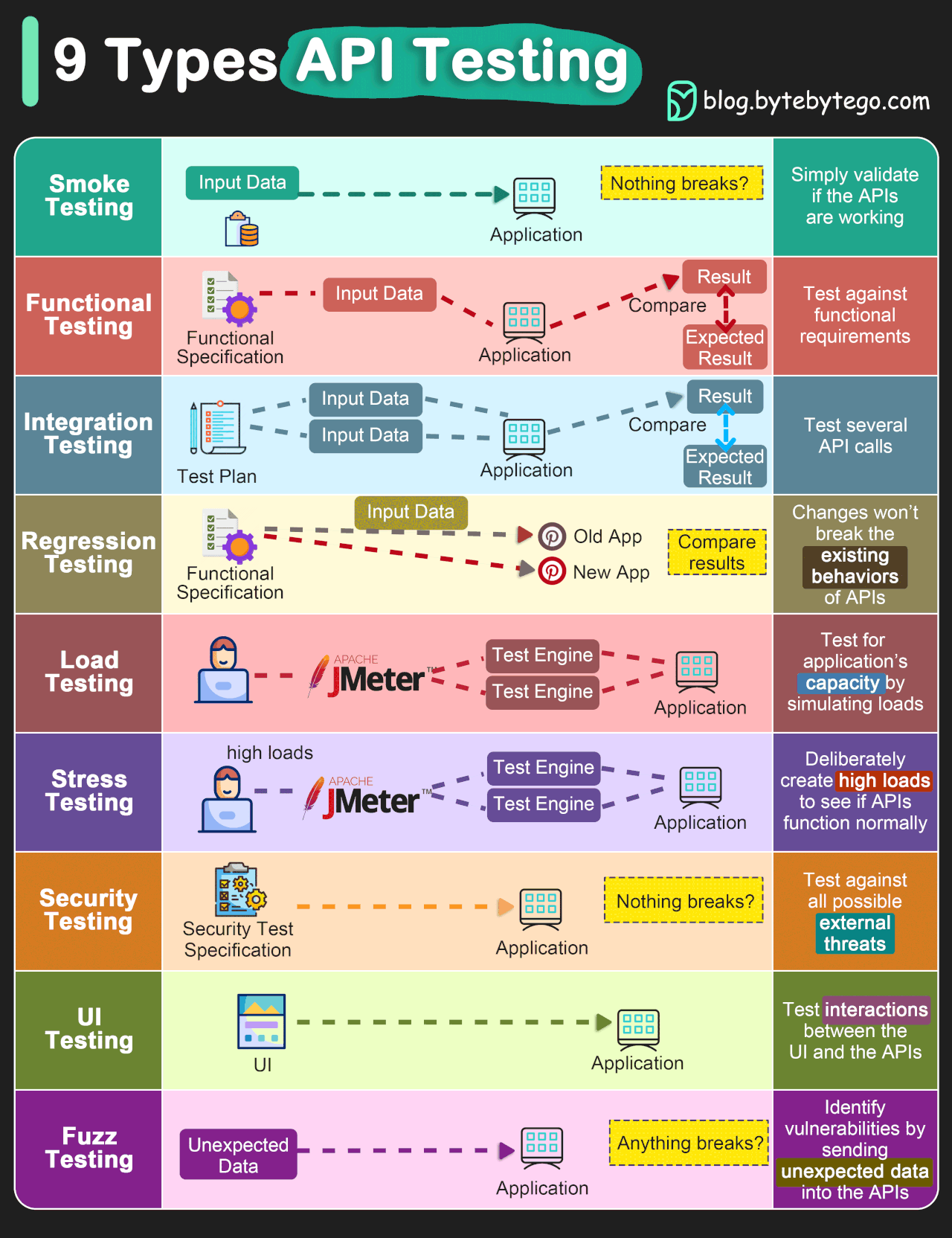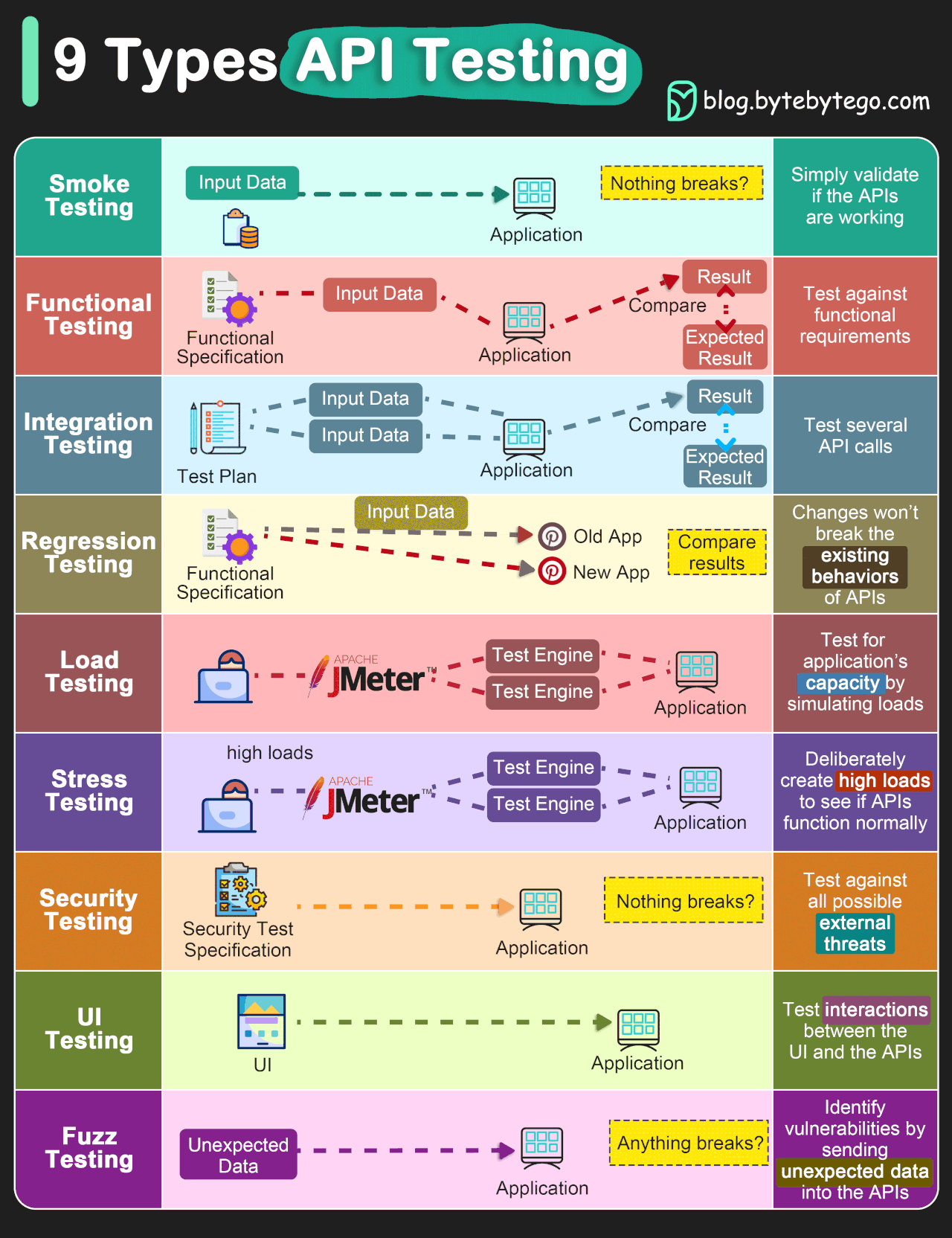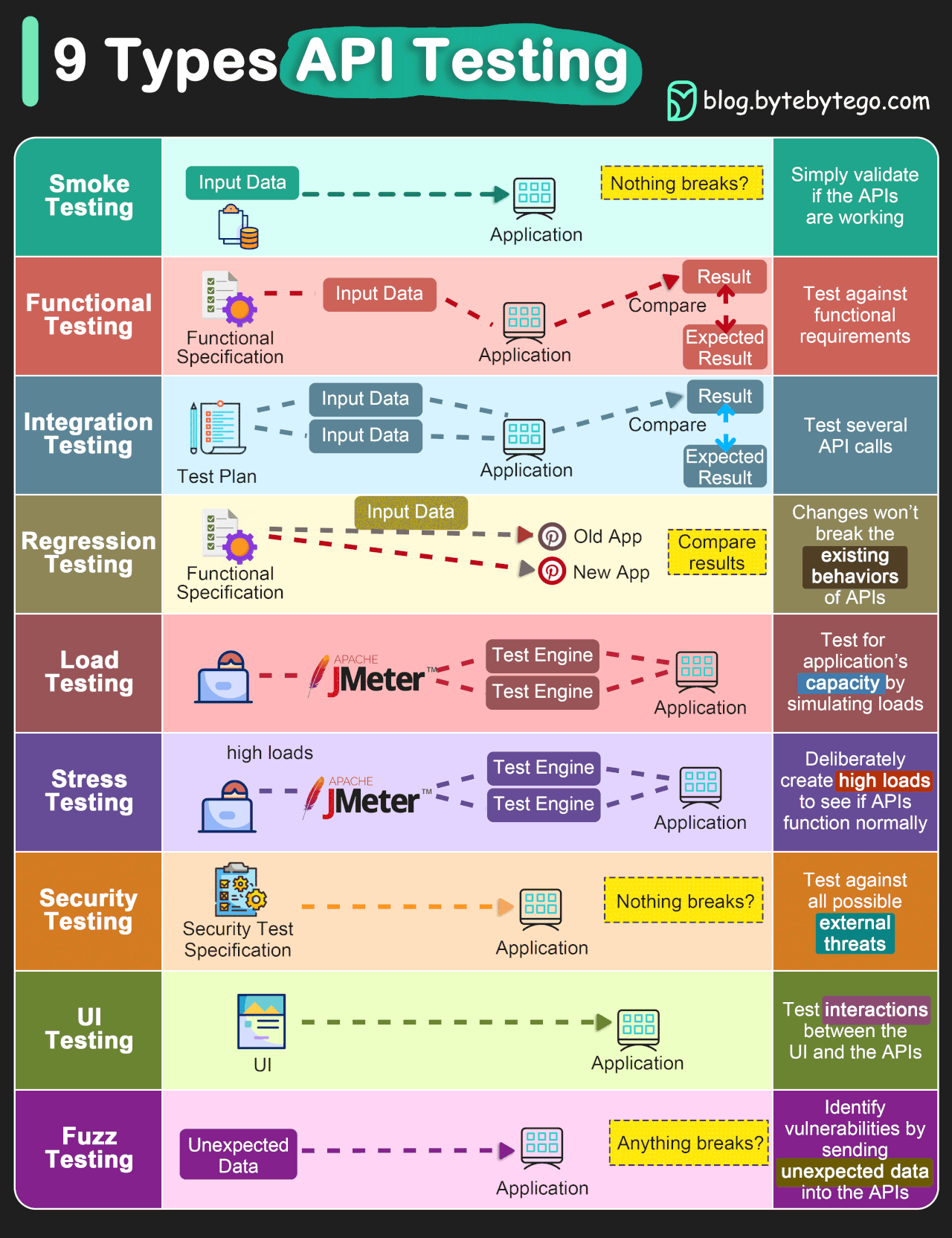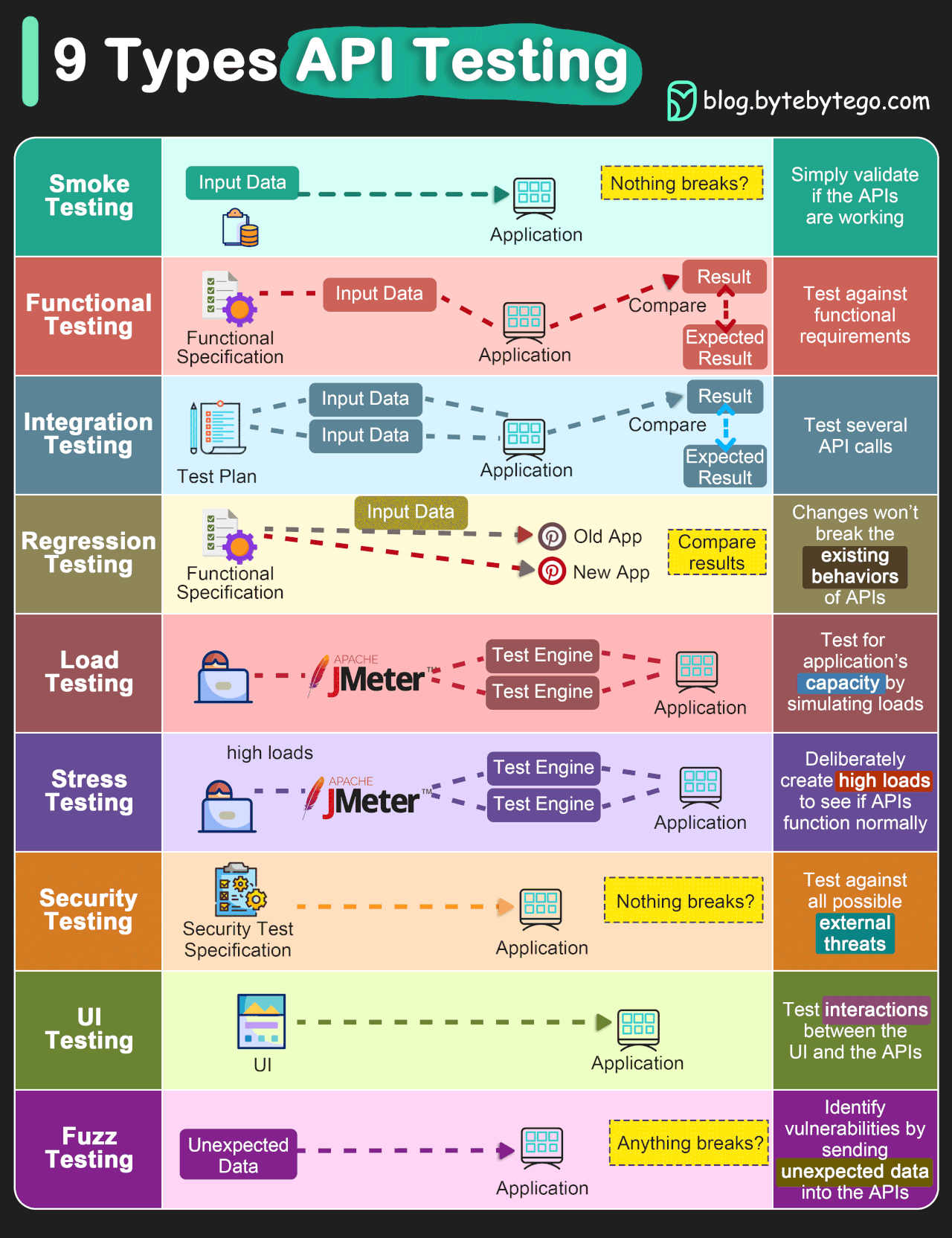
Image credit: ByteByteGo.
However, before we will check out the different test options, we have to talk about one aspect that is central to all: assert.
Assert - a test’s best friend#
In order to check and evaluate if a certain piece of code is doing what it is supposed to do, in a reliable manner, we need a way of asserting what the “correct output” should be and testing the outcome we get against it.
In Python, assert is a statement used to test whether a condition is true. If the condition is true, the program continues to execute as normal. If the condition is false, the program raises an AssertionError exception and optionally can display an accompanying message. The primary use of assert is for debugging and testing purposes, where it helps to catch errors early by ensuring that certain conditions hold at specific points in the code.
Syntax#
The basic syntax of an assert statement is:
assert condition, "Optional error message"
condition: This is the expression to betested. If theconditionevaluates toTrue, nothing happens, and the program continues to execute. If it evaluates toFalse, anAssertionErroris raised."Optional error message": This is the message that is shown when theconditionisfalse. This message is optional, but it’s helpful for understanding why theassertionfailed.
A simple example#
Here’s a simple example demonstrating how assert might be used in a test case:
def add(a, b):
return a + b
# Test case for the add function
def test_add():
result = add(2, 3)
assert result == 5, "Expected add(2, 3) to be 5"
test_add()
If add(2, 3) did not return 5, the assert statement raises an AssertionError with the message "Expected add(2, 3) to be 5".
Usage in Testing#
In the context of testing, assert statements are used to verify that a function or a piece of code behaves as expected. They are a simple yet powerful tool for writing test cases, where you check the outcomes of various functions under different inputs. Here’s how you might use assert in a test:
Checking Function Outputs: To verify that afunctionreturns the expectedvalue.
def add(a, b):
return a + b
# Assert that the function output is as expected
result = add(3, 4)
assert result == 7, f"Expected 7, but got {result}"
result = add(4, 4)
assert result == 7, f"Expected 7, but got {result}"
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
Cell In[4], line 2
1 result = add(4, 4)
----> 2 assert result == 7, f"Expected 7, but got {result}"
AssertionError: Expected 7, but got 8
Validating Data Types: To ensure thatvariablesorreturn valuesare of thecorrect type.
def get_name(name):
return name
# Assert that the return value is a string
name = get_name("Alice")
assert isinstance(name, str), f"Expected type str, but got {type(name).__name__}"
name = get_name(1)
assert isinstance(name, str), f"Expected type str, but got {type(name).__name__}"
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
Cell In[6], line 2
1 name = get_name(1)
----> 2 assert isinstance(name, str), f"Expected type str, but got {type(name).__name__}"
AssertionError: Expected type str, but got int
Testing Invariants: To checkconditionsthat should always betruein a given context.
def add_to_list(item, lst):
lst=[]
lst.append(item)
return lst
# Testing the invariant: the list should not be empty after adding an item
items = add_to_list("apple", [])
assert items, "List should not be empty after adding an item"
Comparing Data Structures: To ensure thatlists,dictionaries,sets, etc., contain the expectedelements.
actual_list = [1, 2, 3]
expected_list = [1, 2, 3]
# Assert that both lists are identical
assert actual_list == expected_list, f"Expected {expected_list}, but got {actual_list}"
actual_list = [1, 2, 4]
expected_list = [1, 2, 3]
# Assert that both lists are identical
assert actual_list == expected_list, f"Expected {expected_list}, but got {actual_list}"
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
Cell In[9], line 5
2 expected_list = [1, 2, 3]
4 # Assert that both lists are identical
----> 5 assert actual_list == expected_list, f"Expected {expected_list}, but got {actual_list}"
AssertionError: Expected [1, 2, 3], but got [1, 2, 4]
Best Practices#
Use for Testing: Leverageassertprimarily intesting frameworksor during thedebugging phase, not as a mechanism for handlingruntime errorsin productioncode.Clear Messages: Includeclear,descriptive messageswithassert statementsto make it easier to identify the cause of atestfailure.Test Precisely: Eachassertshouldtestone specific aspect of yourcode’s behavior to make diagnosing issues straightforward.
Runtime testing#
Runtime tests are tests that run as part of the program itself. They may take the form of checks within the code, as shown below:
For example, we could use the following runtime tests to test the first block of our emd_clust_cont_loss_sim.py script:
# Import all necessary modules and functions
import os
import numpy as np
# Define paths for all to be generated outputs
output_path = "./outputs/"
output_path_data = "./outputs/data"
# For all output paths it should be checked if
# they exist and if not, they should be created
list_paths = [output_path, output_path_data]
# Loop over the list of paths that should be checked
for outputs in list_paths:
# if path exists, all good
if os.path.isdir(outputs):
print(f"{outputs} exists.")
# if not, path should be created
else:
print(f"{outputs} does not exist, creating it.")
os.makedirs(outputs)
# Define seeds to address randomness
np.random.seed(42)
# Create, check and save simulated data
data = np.random.randn(2000, 100)
# Check data shape and if incorrect, raise error
if data.shape==(2000, 100):
print('data shape as expected: %s' %str(data.shape))
else:
print('data shape should be (2000, 100) but got: %s' % str(data.shape))
raise RuntimeError
./outputs/ does not exist, creating it.
./outputs/data does not exist, creating it.
data shape as expected: (2000, 100)
Advantages of runtime testing:#
runwithin the program, so can catch problems caused by logic errors or edge casesmakes it easier to find the cause of the
bugby catching problems earlycatching problems early also helps prevent them escalating into catastrophic failures. It minimises the blast radius.
Disadvantages of runtime testing:#
testscan slow down the programwhat is the right thing to do if an
erroris detected? How should thiserrorbe reported? Exceptions are a recommended route to go with this.
Testing frameworks#
Before if continue to explore different types of testing, we need to talk about testing frameworks as
they are essential in the software development process, enabling developers and researchers to ensure their code behaves as expected. These frameworks facilitate various types of testing, such as unit testing, integration testing, functional testing, regression testing, and performance testing. By automating the execution of tests, verifying outcomes, and reporting results, testing frameworks help improve code quality and software stability.
Importantly, you should start utilizing them right away and don’t switch to a testing framework later on in your project. While you could of course do that, using them from the get go will save you a lot of time and help catch/prevent errors immediately. That’s why we talk about them before addressing different test types so that we can showcase the latter in a test framework for best practice.
Key Features of Testing Frameworks#
Test Organization: Helps structure and manage tests effectively.
Fixture Management: Supports setup and teardown operations for tests.
Assertion Support: Provides tools for verifying test outcomes.
Automated Test Discovery: Automatically identifies and runs tests.
Mocking and Patching: Allows isolation of the system under test.
Parallel Test Execution: Reduces test suite execution time.
Extensibility: Offers customization through plugins and hooks.
Reporting: Generates detailed reports on test outcomes.
Setup for using testing frameworks#
While you could add tests directly to your code, we recommend refactoring and reorganizing your project setup slightly to keep everything organized, testable, and maintainable without duplicating code between your analysis and test scripts.
In general, this entails the following aspects:
have separate scripts for the
code/analysesand its respectivetestswrite
functionsfor parts of yourcodethat run certain tasks/elements (e.g.data downloading,preprocessing,analyses)importandtestthesefunctionsin thetest scripts
With that, you have one “truth” of the code/analyses and can automatically test different parts of them using the testing framework.

There are many different testing frameworks for python, including behave, Robot Framework and TestProject. However, we will focus on pytest.
pytest#
pytest is a powerful testing framework for Python that is easy to start with but also supports complex functional testing. It is known for its simple syntax, detailed assertion introspection, automatic test discovery, and a wide range of plugins and integrations.
Running pytest#
There are various options to run pytest. Let’s start with the easiest one, running all tests written in a specific test directory, ie the setup we introduce above.
At first, you need to ensure pytest is installed in your computational environment. If not, install it using:
%%bash
pip install pytest
Requirement already satisfied: pytest in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (8.3.3)
Requirement already satisfied: pluggy<2,>=1.5 in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (from pytest) (1.5.0)
Requirement already satisfied: iniconfig in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (from pytest) (2.0.0)
Requirement already satisfied: exceptiongroup>=1.0.0rc8 in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (from pytest) (1.2.0)
Requirement already satisfied: tomli>=1 in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (from pytest) (2.0.1)
Requirement already satisfied: packaging in /Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages (from pytest) (23.2)
Additionally, you have to make sure that all your tests are placed in a dedicated directory and that their filenames follows one of these patterns: test_*.py or *_test.py.
they should ideally be placed in a dedicated directory that is different from the path where you work on the code/analysis scripts. Thus, let’s create a respective test directory.
import os
os.makedirs('./tests', exist_ok=True)
Next, we will create our test files and save them in the test directory.
Writing Basic Tests with pytest#
Tests in pytest are simple to write. Starting with test functions, as tests grow, pytest provides a rich set of features for more complex scenarios. The crucial parts as mentioned before are: assert and using functions. You can write one test file per test or have multiple tests in a single test file based on their intended behavior/function.
We will start with a very simple example of the former.
%%writefile ./tests/test_addition.py
def test_example():
assert 1 + 1 == 2
Writing ./tests/test_addition.py
We can now run pytest by pointing it to the directory where we created and saved the test files.
%%bash
pytest tests
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 1 item
tests/test_addition.py::test_example PASSED [100%]
============================== 1 passed in 0.04s ===============================
pytest will automatically discover tests within any files that match the pattern described above in the directory and its subdirectories. As you can see above, it provides a comprehensive output report with details about the computational environment (used software and its version), as well as how many tests were run, their coverage of the code and if they resulted in pass or fail.
Using this setup it’s easy to run automated tests whenever you change something in your code/analyses and evaluate if that had an effect on the expected behavior/function of the code.
Let’s check a few more things about pytest that will come in handy when working with complex scripts.
Using Fixtures for Setup and Teardown#
pytest fixtures define setup and teardown logic for tests, ensuring tests run under controlled conditions.
%%writefile ./tests/test_fixture.py
import pytest
@pytest.fixture
def sample_data():
return [1, 2, 3, 4, 5]
@pytest.mark.sum
def test_sum(sample_data):
assert sum(sample_data) == 15
Writing ./tests/test_fixture.py
Parameterizing Tests#
pytest allows running a single test function with different inputs using @pytest.mark.parametrize.
%%writefile ./tests/test_parameterized.py
import pytest
@pytest.mark.parametrize("a,b,expected", [(1, 1, 2), (2, 3, 5), (3, 3, 6)])
def test_addition(a, b, expected):
assert a + b == expected
Writing ./tests/test_parameterized.py
We can now run all tests by pointing pytest to the respective directory:
%%bash
pytest tests
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 5 items
tests/test_addition.py::test_example PASSED [ 20%]
tests/test_fixture.py::test_sum PASSED [ 40%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 60%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 80%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [100%]
============================== 5 passed in 0.16s ===============================
or run specific tests, e.g. tests from a specific file or those matching a certain pattern.
You can specify the file like so:
%%bash
pytest tests/test_fixture.py
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 1 item
tests/test_fixture.py::test_sum PASSED [100%]
============================== 1 passed in 0.02s ===============================
or run tests matching a name pattern like so:
%%bash
pytest tests/ -k "test_addition"
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 5 items / 1 deselected / 4 selected
tests/test_addition.py::test_example PASSED [ 25%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 50%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 75%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [100%]
======================= 4 passed, 1 deselected in 0.03s ========================
You can also run tests marked with a Custom Marker: If you’ve used custom markers to decorate your tests (e.g., @pytest.mark.regression), you can run only the tests with that marker:
%%bash
pytest tests/ -m "sum" -v
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 5 items / 4 deselected / 1 selected
tests/test_fixture.py::test_sum PASSED [100%]
======================= 1 passed, 4 deselected in 0.02s ========================
NB: we do get a warning related to an unregistered marker as we didn’t create a pytest.ini file yet.
The pytest.ini file is a configuration file for pytest that allows you to specify settings, register custom markers, configure test paths, and more. It is typically placed in the root directory of your project and helps streamline the pytest setup by keeping configurations in one place.
Thus, let’s create one.
%%writefile ./pytest.ini
[pytest]
# Register custom markers to avoid warnings
markers =
sum: mark a test as related to summation operations
# Default command-line options
addopts = -v
# Specify default test directories
testpaths = ./tests
# Specify the path for the source files
pythonpath = .
Overwriting ./pytest.ini
With that in place, we can now run pytest with even less effort.
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 5 items
tests/test_addition.py::test_example PASSED [ 20%]
tests/test_fixture.py::test_sum PASSED [ 40%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 60%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 80%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [100%]
============================== 5 passed in 0.02s ===============================
After learning about the basics of testing frameworks, it’s time to check how we can use them to implement different test types by also incorporating the setup we talked about before.
Code testing - different types of tests#
We already explored different types of tests a little bit in the above section but it’s time to go more into detail and learned more about common test types by using them for our code/analyses.
Smoke tests#
These are very brief initial checks that ensures the basic requirements required to run the code hold. If these fail there is no point proceeding to additional levels of testing until they are fixed.
One example for our code/analyses would be to check if the directories in the output paths are created, as without them, we wouldn’t get our outputs saved at all or somewhere else.
As mentioned before, to follow best practices and make things manageable and easy for the long run, we need to refractor our code/analyses to run certain parts within functions that we can import in the tests.
For the output directory generation part, we would change things from:
# Import all necessary modules and functions
import os
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
from sklearn.mixture import BayesianGaussianMixture
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
from joblib import dump
import matplotlib.pyplot as plt
import seaborn as sns
# Define paths for all to be generated outputs
output_path = "./outputs/"
output_path_data = "./outputs/data"
output_path_graphics = "./outputs/graphics"
output_path_models = "./outputs/models"
# For all output paths it should be checked if
# they exist and if not, they should be created
list_paths = [output_path, output_path_data, output_path_graphics,
output_path_models]
# Loop over the list of paths that should be checked
for outputs in list_paths:
# if path exists, all good
if os.path.isdir(outputs):
print(f"{outputs} exists.")
# if not, path should be created
else:
print(f"{outputs} does not exist, creating it.")
os.makedirs(outputs)
to
# Import all necessary modules
import os
# Define utility function to create paths
def create_output_paths(paths):
for path in paths:
if not os.path.isdir(path):
os.makedirs(path)
return paths
# Run all steps of the code/analysis
def main():
# Set up output directories
paths = ["./outputs/", "./outputs/data", "./outputs/graphics", "./outputs/models"]
create_output_paths(paths)
if __name__ == "__main__":
main()
So, let’s create a respective version of our code/analyses to make this possible. It’s not best practice but for now we will create a new file instead of updating the old one so that we can easily compare them.
%%writefile ./emd_clust_cont_loss_sim_functions.py
# Import all necessary modules
import os
# Define utility function to create paths
def create_output_paths(paths):
for path in paths:
if not os.path.isdir(path):
os.makedirs(path)
return paths
# Define test directories
paths = ["./outputs/", "./outputs/data", "./outputs/graphics", "./outputs/models"]
# Run all steps of the code/analysis
def main():
# Set up output directories
create_output_paths(paths)
if __name__ == "__main__":
main()
Overwriting ./emd_clust_cont_loss_sim_functions.py
We can now setup a smoke test to evaluate if the output paths are generated as expected by utilizing, ie importing, the just defined function.
%%writefile ./tests/test_paths.py
import os
import shutil
import pytest
from emd_clust_cont_loss_sim_functions import create_output_paths
# Define test paths (use temporary or unique paths to avoid overwriting real data)
test_paths = ["./test_outputs/", "./test_outputs/data", "./test_outputs/graphics", "./test_outputs/models"]
# Define tests
def test_create_output_paths():
# Run the path creation function
create_output_paths(test_paths)
# Check if each path now exists
for path in test_paths:
assert os.path.isdir(path), f"Expected directory {path} to be created, but it was not."
# Cleanup - remove the directories after the test to keep the environment clean
for path in reversed(test_paths): # Remove subdirectories before parent directory
if os.path.isdir(path):
os.rmdir(path)
# Cleanup - remove generated outputs to keep the test environment clean
for path in reversed(test_paths):
if os.path.isdir(path):
shutil.rmtree(path)
Writing ./tests/test_paths.py
With everything in place, we can now run pytest as we did before, pointing it to the specific test that we would like to run:
%%bash
pytest tests/test_paths.py
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
plugins: anyio-4.2.0
collecting ... collected 1 item
tests/test_paths.py::test_create_output_paths PASSED [100%]
============================== 1 passed in 0.03s ===============================
That appears to work out, great! We could also run all tests like so:
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 6 items
tests/test_addition.py::test_example PASSED [ 16%]
tests/test_fixture.py::test_sum PASSED [ 33%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 50%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 66%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [ 83%]
tests/test_paths.py::test_create_output_paths PASSED [100%]
============================== 6 passed in 0.04s ===============================
Isn’t that great? Let’s continue with exploring and adding more test types!
Unit tests#
A level of the software testing process where individual units of a software are tested. The purpose is to validate that each unit of the software performs as designed. For example, in our code/analyses, we could evaluate if the functions for path and data generation work as expected. Thus, we could write a unit test for both comparing the expected to the actual output.
At first, we need to refactor the data generation in our code/analyses to be run as a function, ie adding it to the new version of the script.
Overwriting ./emd_clust_cont_loss_sim_functions.py
Subsequently, we need to write respective unit tests.
%%writefile ./tests/test_path_sim_data_gen.py
import os
import shutil
import numpy as np
import pytest
from emd_clust_cont_loss_sim_functions import create_output_paths, simulate_data
# Define temporary test paths to avoid overwriting real data
test_paths = ["./test_outputs/", "./test_outputs/data", "./test_outputs/graphics", "./test_outputs/models"]
def test_create_output_paths():
# Run the path creation function
create_output_paths(test_paths)
# Check if each path now exists
for path in test_paths:
assert os.path.isdir(path), f"Expected directory {path} to be created, but it was not."
def test_simulate_data():
# Generate test data
data = simulate_data(size=(100, 50), save_path="./test_outputs/data/raw_data_sim.npy")
# Check type and shape
assert isinstance(data, np.ndarray), "Expected output type np.ndarray"
assert data.shape == (100, 50), "Expected output shape (100, 50)"
os.remove("./test_outputs/data/raw_data_sim.npy")
# Cleanup - remove generated outputs to keep the test environment clean
for path in reversed(test_paths):
if os.path.isdir(path):
shutil.rmtree(path)
Writing ./tests/test_path_sim_data_gen.py
Using our, by now, well-know pytest, we run the unit tests.
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 8 items
tests/test_addition.py::test_example PASSED [ 12%]
tests/test_fixture.py::test_sum PASSED [ 25%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 37%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 50%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [ 62%]
tests/test_path_sim_data_gen.py::test_create_output_paths PASSED [ 75%]
tests/test_path_sim_data_gen.py::test_simulate_data PASSED [ 87%]
tests/test_paths.py::test_create_output_paths PASSED [100%]
============================== 8 passed in 0.18s ===============================
Unit Testing Tips#
many
testing frameworkshavetoolsspecifically geared towards writing and running unit tests,pytestdoes as well
isolate the
development environmentfrom thetest environment
write
test casesthat are independent of each other. For example, if aunit Autilises the result supplied by anotherunit B, you shouldtestunit Awith atest double, rather than actually calling theunit B. If you don’t do this yourtestfailingmay be due to a fault in eitherunit Aorunit B, making thebugharder to trace.
aim at
coveringallpathsthrough aunit, pay particular attention toloop conditions.
in addition to
writing casesto verify the behaviour, writecasesto ensure theperformanceof thecode. For example, if afunctionthat is supposed to addtwo numberstakes several minutes to run there is likely a problem.
if you find a defect in your
codewrite atestthat exposes it. Why? First, you will later be able to catch the defect if you do not fix it properly. Second, yourtest suiteis now more comprehensive. Third, you will most probably be too lazy to write thetestafter you have already fixed the defect.
Integration tests#
A level of software testing where individual units are combined and tested as a group. The purpose of this level of testing is to expose faults in the interaction between integrated units.
Integration Testing Approaches#
There are several different approaches to integration testing.
Big Bang: an approach tointegration testingwhere all or most of theunitsare combined together and tested at one go. This approach is taken when thetesting teamreceives the entiresoftwarein a bundle. So what is the difference betweenBig Bangintegration testingandsystem testing? Well, the formertestsonly theinteractionsbetween theunitswhile the latterteststhe entire system.
Top Down: an approach tointegration testingwheretop-level sectionsof thecode(that themselves contain many smallerunits) aretested firstandlower level unitsaretestedstep by step after that.
Bottom Up: an approach tointegration testingwhereintegrationbetween bottom level sections are tested first and upper-level sections step by step after that. Againtest stubsshould be used, in this case tosimulateinputsfrom higher level sections.
Sandwich/Hybridis an approach tointegration testingwhich is a combination ofTop DownandBottom Upapproaches.
Which approach you should use will depend on which best suits the nature/structure of your project.
Based on our code/analyses, we could test a sequence of steps from data generation, model training, and output generation.
End-to-End PathandDirectoryCreation: Verify thatcreate_output_pathssets up all necessarydirectories, and thesedirectoriesare populated withfilesordatagenerated by the pipeline.Data GenerationtoModel Training:Simulate data,splitit, and ensure that themodelcan betrainedon thisdata.Model TrainingwithOutput Verification: Run a shorttrainingsession and verify that thelossdecreases over a fewepochs, indicating themodelislearning.
The first step, as always, is to adapt our code/analyses so that all needed aspects are run in via functions.
Overwriting ./emd_clust_cont_loss_sim_functions.py
Next, we need to write tests that evaluate the included parts of the code/analyses back-to-back.
%%writefile ./tests/test_pipeline.py
# Import all necessary modules
import os
import shutil
import numpy as np
import torch
from emd_clust_cont_loss_sim_functions import create_output_paths, simulate_data, Encoder, train_encoder
# Define list of paths
test_paths = ["./test_outputs/", "./test_outputs/data", "./test_outputs/graphics", "./test_outputs/models"]
# Define the tests
def test_end_to_end_pipeline():
# Step 1: Set up output directories
create_output_paths(test_paths)
for path in test_paths:
assert os.path.isdir(path), f"Directory {path} was not created."
# Step 2: Generate data
data = simulate_data(size=(100, 50), save_path="./test_outputs/data/raw_data_sim.npy")
assert isinstance(data, np.ndarray), "Expected output type np.ndarray"
assert data.shape == (100, 50), "Expected output shape (100, 50)"
# Step 3: Split data and prepare for model training
data_train = torch.from_numpy(data[:80]).float()
# Step 4: Initialize and train the Encoder model for a few epochs
encoder = Encoder(input_dim=50, hidden_dim=25, embedding_dim=10)
losses, _ = train_encoder(encoder, data_train, epochs=10) # Limit epochs for a quick test
# Check that the losses are recorded and decrease over epochs
assert len(losses) == 10, "Expected 10 loss values, one for each epoch."
assert losses[-1] < losses[0], "Expected the loss to decrease over training."
# Cleanup - remove generated outputs to keep the test environment clean
for path in reversed(test_paths):
if os.path.isdir(path):
shutil.rmtree(path)
Writing ./tests/test_pipeline.py
With that , we are already back to running out tests, this time including a rather complex and holistic integration test. Please note, that tests might start taking longer, as we actually running, ie testing quite a bite of code, including model training and evaluation.
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 9 items
tests/test_addition.py::test_example PASSED [ 11%]
tests/test_fixture.py::test_sum PASSED [ 22%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 33%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 44%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [ 55%]
tests/test_path_sim_data_gen.py::test_create_output_paths PASSED [ 66%]
tests/test_path_sim_data_gen.py::test_simulate_data PASSED [ 77%]
tests/test_paths.py::test_create_output_paths PASSED [ 88%]
tests/test_pipeline.py::test_end_to_end_pipeline PASSED [100%]
============================== 9 passed in 4.33s ===============================
Integration Testing Tips#
Ensure that you have a proper Detail Design document where interactions between each unit are clearly defined. It is difficult or impossible to perform integration testing without this information.
Make sure that each unit is unit tested and fix any bugs before you start integration testing. If there is a bug in the individual units then the integration tests will almost certainly fail even if there is no error in how they are integrated.
Use mocking/stubs where appropriate.
System tests#
A level of the software testing process where a complete, integrated system is tested. The purpose of this test is to evaluate whether the system as a whole gives the correct outputs for given inputs.
System Testing Tips#
System tests, also called end-to-end tests, run the program, well, from end to end. As such these are the most time consuming tests to run. Therefore you should only run these if all the lower-level tests (smoke, unit, integration) have already passed. If they haven’t, fix the issues they have detected first before wasting time running system tests.
Because of their time-consuming nature it will also often be impractical to have enough system tests to trace every possible route through a program, especially if there are a significant number of conditional statements. Therefore you should consider the system test cases you run carefully and prioritise:
the most common routes through a program
the most important routes for a program
casesthat are prone to breakage due to structural problems within the program. Though ideally it’s better to justfixthose problems, butcasesexist where this may not be feasible.
Because system tests can be time consuming it may be impractical to run them very regularly (such as multiple times a day after small changes in the code). Therefore it can be a good idea to run them each night (and to automate this process) so that if errors are introduced that only system testing can detect, the developer(s) will be made aware of them relatively quickly.
In order to run a system test for our code/analyses, we need to refactor the entire script to utilize functions.
Overwriting ./emd_clust_cont_loss_sim_functions.py
Now to the second part: writing a system level test. Here’s what it should do:
The
scriptrunsend-to-endwithout errors, simulating a full workflow execution.Output directoriesandfilesare created as expected, confirming each step’s success.Modelbehavior is evaluated by checking if it can produceembeddingsand iftraining resultsin areductionofloss.
%%writefile ./tests/test_system_analyses.py
import os
import shutil
import numpy as np
import torch
from emd_clust_cont_loss_sim_functions import main, Encoder, simulate_data, train_encoder
def test_system_workflow():
# Set up paths for verification
output_paths = ["./outputs/", "./outputs/data", "./outputs/graphics", "./outputs/models"]
data_file = "./outputs/data/raw_data_sim.npy"
plot_file = "./outputs/graphics/data_examples.png"
model_file = "./outputs/models/encoder.pth"
# Run the main function to execute the workflow
main()
# Check that output directories are created
for path in output_paths:
assert os.path.isdir(path), f"Expected directory {path} to be created."
# Verify data file existence and format
assert os.path.isfile(data_file), "Expected data file not found."
data = np.load(data_file)
assert isinstance(data, np.ndarray), "Data file is not in the expected .npy format."
# Verify the plot file is created
assert os.path.isfile(plot_file), "Expected plot file not found."
# Verify the model file is created
assert os.path.isfile(model_file), "Expected model file not found."
# Step 1: Load and check model
encoder = torch.load(model_file)
assert isinstance(encoder, Encoder), "The loaded model is not an instance of the Encoder class."
# Step 2: Evaluate model with sample data
# Use a small subset of the data to generate embeddings and verify the output shape
data_sample = torch.from_numpy(data[:10]).float() # Take a sample of 10 for testing
with torch.no_grad():
embeddings = encoder(data_sample)
# Check the output embeddings shape
expected_shape = (10, encoder.layers[-1].out_features)
assert embeddings.shape == expected_shape, f"Expected embeddings shape {expected_shape}, but got {embeddings.shape}."
# Step 3: Check if training reduces loss over epochs
# Train the model for a small number of epochs and confirm that loss decreases
data_train = torch.from_numpy(data[:80]).float()
initial_loss, final_loss = None, None
encoder = Encoder(input_dim=100, hidden_dim=100, embedding_dim=50) # Reinitialize the encoder for testing
losses, _ = train_encoder(encoder, data_train, epochs=10) # Train for a few epochs for quick testing
# Capture initial and final losses
initial_loss, final_loss = losses[0], losses[-1]
# Verify that the final loss is lower than the initial loss
assert final_loss < initial_loss, "Expected final loss to be lower than initial loss, indicating training progress."
# Cleanup - remove generated outputs to keep the test environment clean
for path in reversed(output_paths):
if os.path.isdir(path):
shutil.rmtree(path)
Writing ./tests/test_system_analyses.py
The moment of truth is here: we run the tests as usual and will grab something to drink/snack, as this might take a while.
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 10 items
tests/test_addition.py::test_example PASSED [ 10%]
tests/test_fixture.py::test_sum PASSED [ 20%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 30%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 40%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [ 50%]
tests/test_path_sim_data_gen.py::test_create_output_paths PASSED [ 60%]
tests/test_path_sim_data_gen.py::test_simulate_data PASSED [ 70%]
tests/test_paths.py::test_create_output_paths PASSED [ 80%]
tests/test_pipeline.py::test_end_to_end_pipeline PASSED [ 90%]
tests/test_system_analyses.py::test_system_workflow PASSED [100%]
=============================== warnings summary ===============================
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/utils.py:11
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/utils.py:11: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import pandas as pd
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/rcmod.py:82
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/rcmod.py:82: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
if LooseVersion(mpl.__version__) >= "3.0":
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/setuptools/_distutils/version.py:346
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/setuptools/_distutils/version.py:346: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
other = LooseVersion(other)
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582: MatplotlibDeprecationWarning: The register_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps.register(name)`` instead.
mpl_cm.register_cmap(_name, _cmap)
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583: MatplotlibDeprecationWarning: The register_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps.register(name)`` instead.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
tests/test_system_analyses.py::test_system_workflow
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
tests/test_system_analyses.py::test_system_workflow
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/threadpoolctl.py:1010: RuntimeWarning:
Found Intel OpenMP ('libiomp') and LLVM OpenMP ('libomp') loaded at
the same time. Both libraries are known to be incompatible and this
can cause random crashes or deadlocks on Linux when loaded in the
same Python program.
Using threadpoolctl may cause crashes or deadlocks. For more
information and possible workarounds, please see
https://github.com/joblib/threadpoolctl/blob/master/multiple_openmp.md
warnings.warn(msg, RuntimeWarning)
-- Docs: https://docs.pytest.org/en/stable/how-to/capture-warnings.html
======================= 10 passed, 17 warnings in 16.15s =======================
Acceptance and regression tests#
A level of the software testing process where a system is tested for acceptability. The purpose of this test is to evaluate the system’s compliance with the project requirements and assess whether it is acceptable for the purpose.
Acceptance testing#
Acceptance tests are one of the last tests types that are performed on software prior to delivery. Acceptance testing is used to determine whether a piece of software satisfies all of the requirements from user’s perspective. Does this piece of software do what it needs to do? These tests are sometimes built against the original specification.
Because research software is typically written by the researcher that will use it (or at least with significant input from them) acceptance tests may not be necessary.
Regression testing#
Regression testing checks for unintended changes by comparing new test results to previous ones, ensuring updates don’t break the software. It’s critical because even unrelated code changes can cause issues. Suitable for all testing levels, it’s vital in system testing and can automate tedious manual checks. Tests are created by recording outputs for specific inputs, then retesting and comparing results to detect discrepancies. Essential for team projects, it’s also crucial for solo work to catch self-introduced errors.
Regression testing approaches differ in their focus. Common examples include:
Bug regression: retest a specificbugthat has been allegedlyfixed
Old fix regression testing: retest several oldbugsthat werefixed, to see if they are back. (This is the classical notion ofregression: the program has regressed to a bad state.)
General functional regression:retestthe project broadly, including areas that worked before, to see whether more recent changes have destabilized workingcode.
Conversion or port testing: the program isportedto a new platform and aregression test suiteis run to determine whether theportwas successful.
Configuration testing: theprogramisrunwith a newdeviceor on a newversionof theoperating systemor in conjunction with a new application. This is likeport testingexcept that the underlyingcodehasn’t been changed–only the externalcomponentsthat thesoftwareundertestmust interact with.
As we already refactored the entire code/analyses to utilize ``functions, we can start writing a set of regression testsright away. In more detail we will ensure that the generateddata, model training, and model output` remain consistent:
Check Data Consistency: Verify that thedatagenerated bysimulate_datahas aspecific shapeandsample values.Check Loss Consistency:TraintheEncoderfor a fewepochsand confirm that the finallossfalls within anexpected range, ensuring themodeltrainsas expected.Check Model Output Consistency: Confirm that the outputembeddingshave the correctshapeand that specificvaluesin the output are as expected.
%%writefile ./tests/test_regression_analyses.py
# Import all necessary modules
import os
import shutil
import numpy as np
import torch
from emd_clust_cont_loss_sim_functions import simulate_data, Encoder, train_encoder
# Define list of paths
test_paths = ["./test_outputs/", "./test_outputs/data", "./test_outputs/graphics", "./test_outputs/models"]
# Define regression test
def test_regression_workflow():
# Part 1: Data Consistency Check
# Generate test data and check shape and sample values
data = simulate_data(size=(100, 100), save_path="./test_outputs/data/raw_data_sim.npy")
# Verify data shape
assert data.shape == (100, 100), "Data shape mismatch."
# Part 2: Loss Consistency Check
# Initialize the encoder and train it briefly, then check final loss
input_dim = 100
encoder = Encoder(input_dim=input_dim, hidden_dim=100, embedding_dim=50)
data_train = torch.from_numpy(data).float()
# Train the encoder for a few epochs
losses, _ = train_encoder(encoder, data_train, epochs=10)
# Check that the final loss is within the expected range
final_loss = losses[-1]
assert 0.01 < final_loss < 2.0, f"Final loss {final_loss} is outside the expected range (0.01, 2.0)."
# Part 3: Model Output Consistency Check
# Generate embeddings and verify their shape and sample values
data_sample = torch.from_numpy(data[:10]).float() # Take a sample of 10 rows for testing
with torch.no_grad():
embeddings = encoder(data_sample)
# Verify output shape
assert embeddings.shape == (10, 50), "Embedding shape mismatch."
# Expected embedding sample values (replace with known values for regression testing)
expected_embedding_sample = embeddings[0].numpy()[:5] # Capture this once and use it as reference
np.testing.assert_almost_equal(embeddings[0].numpy()[:5], expected_embedding_sample, decimal=6,
err_msg="Embedding does not match expected sample values.")
# Cleanup - remove generated outputs to keep the test environment clean
for path in reversed(test_paths):
if os.path.isdir(path):
shutil.rmtree(path)
Writing ./tests/test_regeression_analyses.py
For the last time in this session, let’s run the tests.
%%bash
pytest
============================= test session starts ==============================
platform darwin -- Python 3.10.6, pytest-8.3.3, pluggy-1.5.0 -- /Users/peerherholz/anaconda3/envs/course_name/bin/python3.1
cachedir: .pytest_cache
rootdir: /Users/peerherholz/google_drive/GitHub/Clean_Repro_Code_Neuromatch/workshop/materials/code_form_test_CI
configfile: pytest.ini
testpaths: ./tests
plugins: anyio-4.2.0
collecting ... collected 11 items
tests/test_addition.py::test_example PASSED [ 9%]
tests/test_fixture.py::test_sum PASSED [ 18%]
tests/test_parameterized.py::test_addition[1-1-2] PASSED [ 27%]
tests/test_parameterized.py::test_addition[2-3-5] PASSED [ 36%]
tests/test_parameterized.py::test_addition[3-3-6] PASSED [ 45%]
tests/test_path_sim_data_gen.py::test_create_output_paths PASSED [ 54%]
tests/test_path_sim_data_gen.py::test_simulate_data PASSED [ 63%]
tests/test_paths.py::test_create_output_paths PASSED [ 72%]
tests/test_pipeline.py::test_end_to_end_pipeline PASSED [ 81%]
tests/test_regeression_analyses.py::test_regression_workflow PASSED [ 90%]
tests/test_system_analyses.py::test_system_workflow PASSED [100%]
=============================== warnings summary ===============================
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/utils.py:11
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/utils.py:11: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import pandas as pd
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/rcmod.py:82
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/rcmod.py:82: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
if LooseVersion(mpl.__version__) >= "3.0":
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/setuptools/_distutils/version.py:346
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/setuptools/_distutils/version.py:346: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
other = LooseVersion(other)
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1582: MatplotlibDeprecationWarning: The register_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps.register(name)`` instead.
mpl_cm.register_cmap(_name, _cmap)
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
../../../../../../anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/cm.py:1583: MatplotlibDeprecationWarning: The register_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps.register(name)`` instead.
mpl_cm.register_cmap(_name + "_r", _cmap_r)
tests/test_system_analyses.py::test_system_workflow
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
tests/test_system_analyses.py::test_system_workflow
/Users/peerherholz/anaconda3/envs/course_name/lib/python3.10/site-packages/threadpoolctl.py:1010: RuntimeWarning:
Found Intel OpenMP ('libiomp') and LLVM OpenMP ('libomp') loaded at
the same time. Both libraries are known to be incompatible and this
can cause random crashes or deadlocks on Linux when loaded in the
same Python program.
Using threadpoolctl may cause crashes or deadlocks. For more
information and possible workarounds, please see
https://github.com/joblib/threadpoolctl/blob/master/multiple_openmp.md
warnings.warn(msg, RuntimeWarning)
-- Docs: https://docs.pytest.org/en/stable/how-to/capture-warnings.html
======================= 11 passed, 17 warnings in 16.76s =======================
Conclusion:#
Embracing testing and testing frameworks like pytest and incorporating a comprehensive testing strategy are essential steps towards achieving high-quality software development. These frameworks not only automate the testing process but also provide a structured approach to addressing a wide spectrum of testing requirements. By leveraging their capabilities, researchers and software developers can ensure thorough test coverage, streamline debugging, and maintain high standards of software quality and `performance``.
Task for y’all!
Remember our script from the beginning? You already went through it a couple of times and brought to code (get it?). Now, we would like to add some tests for our script to ensure its functionality.
You have 40 min.
