Note
Go to the end to download the full example code.
Fetching BIDS-Atlas compliant atlases¶
This example demonstrates how to use bids_atlas.datasets to fetch
atlases that confirm to BIDS-Atlas.
Much of the functionality of the bids_atlas toolbox relies on downloading
commonly used publicly available atlases. Each atlas has its own function, with certain
arguments being shared across all of them.
This specifically refers to the target space and resolution the given atlas should
be obtained in.
Here we show how download a few atlases, using the respective functions and arguments.
First of all, we are going to import bids_atlas dataset module, as this will give us
access to all respective functions.
from bids_atlas import datasets
import pandas as pd
from nilearn.plotting import plot_roi
Lets start with the AAL atlas. In order to obtain it in a BIDS-Atlas compliant manner, we only need to
use the respective function, called get_AAL. If we run it without specifying any arguments, it will be provided
in the current directory and default specifications, ie 2mm resolution. The function will return a dictionary with
the paths to atlas image, .tsv and .json files.
AAL_atlas = datasets.get_AAL()
[fetch_atlas_aal] Added README.md to /home/runner/nilearn_data
[fetch_atlas_aal] Dataset created in /home/runner/nilearn_data/aal_3v2
[fetch_atlas_aal] Downloading data from
https://www.gin.cnrs.fr/wp-content/uploads/AAL3v2_for_SPM12.tar.gz ...
[fetch_atlas_aal] ...done. (1 seconds, 0 min)
[fetch_atlas_aal] Extracting data from
/home/runner/nilearn_data/aal_3v2/43f38da73bc7adb6022df5794d84f2eb/AAL3v2_for_SP
M12.tar.gz...
[fetch_atlas_aal] .. done.
Downloading https://templateflow.s3.amazonaws.com/tpl-MNIColin27/tpl-MNIColin27_T1w.nii.gz
0%| | 0.00/14.7M [00:00<?, ?B/s]
56%|█████▌ | 8.28M/14.7M [00:00<00:00, 82.8MB/s]
100%|██████████| 14.7M/14.7M [00:00<00:00, 95.4MB/s]
checking if atlas needs to be resampled
atlas will be resampled to target
/home/runner/work/bids_atlas/bids_atlas/bids_atlas/utils.py:110: FutureWarning: 'force_resample' will be set to 'True' by default in Nilearn 0.13.0.
Use 'force_resample=True' to suppress this warning.
atlas_resampled = resample_to_img(atlas, target, interpolation='nearest')
/home/runner/work/bids_atlas/bids_atlas/bids_atlas/utils.py:110: FutureWarning: From release 0.13.0 onwards, this function will, by default, copy the header of the input image to the output. Currently, the header is reset to the default Nifti1Header. To suppress this warning and use the new behavior, set `copy_header=True`.
atlas_resampled = resample_to_img(atlas, target, interpolation='nearest')
Atlas will be saved to /home/runner/work/bids_atlas/bids_atlas/examples/bids_atlas_datasets/atlas-AAL
Downloaded template metadata to /home/runner/work/bids_atlas/bids_atlas/examples/bids_atlas_datasets/atlas-AAL/tpl-MNIColin27/anat/tpl-MNIColin27.json
The following files were downloaded at /home/runner/work/bids_atlas/bids_atlas/examples/bids_atlas_datasets/atlas-AAL
atlas-AAL/
├─atlas-AAL_description.json
└─tpl-MNIColin27/
└─anat/
├─tpl-MNIColin27_atlas-AAL_res-1_dseg.tsv
├─tpl-MNIColin27.json
├─tpl-MNIColin27_T1w.nii.gz
└─tpl-MNIColin27_atlas-AAL_res-1_dseg.nii.gz
Now the respective files can be accessed via their keys. The path to the atlas image can be obtained via
AAL_atlas['AtlasImage']
'/home/runner/work/bids_atlas/bids_atlas/examples/bids_atlas_datasets/atlas-AAL/tpl-MNIColin27/anat/tpl-MNIColin27_atlas-AAL_res-1_dseg.nii.gz'
and thus easily be loaded, plotted, or utilized within an analysis.
plot_roi(AAL_atlas['AtlasImage'], draw_cross=False, cmap='Set2')
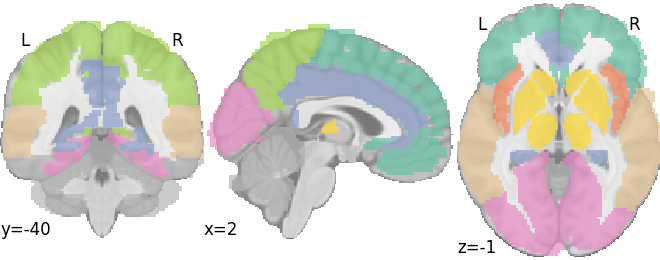
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f2704768e10>
The .tsv and .json files contain important information and metadata concerning the atlas. The former entails a DataFrame indicating the indices of the atlas and details thereof.
pd.read_csv(AAL_atlas['AtlasTSV'], sep='\t')
The latter comprises the atlas’ metadata following BIDS specifications.
import json
with open(AAL_atlas['AtlasJson'], 'r') as AAL_atlas_json:
AAL_atlas_json_load = json.load(AAL_atlas_json)
AAL_atlas_json_load = json.dumps(AAL_atlas_json_load, indent=4)
print(AAL_atlas_json_load)
{
"Name": "Automated Anatomical Labeling Atlas - SPM12, 3v2 version",
"Description": "AAl atlas for SPM 12, 3v2 version. Notes: This atlas is the result of an automated anatomical parcellation\nof the spatially normalized single-subject high-resolution T1 volume provided by the\nMontreal Neurological Institute (MNI) (D. L. Collins et al., 1998, Trans. Med. Imag. 17, 463-468, PubMed).\nUsing this parcellation method, three procedures to perform the automated anatomical labeling of functional\nstudies are proposed: (1) labeling of an extremum defined by a set of coordinates, (2) percentage of voxels belonging\nto each of the AVOI intersected by a sphere centered by a set of coordinates,\nand (3) percentage of voxels belonging to each of the AVOI intersected by an activated cluster.",
"BIDSVersion": "PLEASE ADD",
"Authors": "PLEASE ADD",
"HowToAcknowledge": "PLEASE ADD",
"SourceDatasetsURLs": "PLEASE ADD",
"License": "Unknown",
"Funding": "PLEASE ADD",
"ReferencesAndLinks": "http://www.gin.cnrs.fr/AAL-217?lang",
"Species": "Homo sapiens",
"DerivedFrom": "PLEASE ADD",
"SampleSize": "1",
"SpecialReference": "PLEASE ADD"
}
Total running time of the script: (0 minutes 4.721 seconds)