Gradient profiling
Contents
Gradient profiling#
Import modules#
from copy import deepcopy
import os
import json
from neuromaps import datasets, transforms, images, compare_images, nulls
import nibabel as nb
import numpy as np
from scipy import stats
from brainsmash.mapgen.memmap import txt2memmap
from brainsmash.mapgen.sampled import Sampled
from brainsmash import workbench
from brainsmash.mapgen.eval import sampled_fit
from brainsmash.mapgen.stats import pearsonr, pairwise_r, nonparp
from neuromaps.plotting import plot_surf_template
from nilearn.plotting import view_surf, plot_matrix
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
import cmasher as cmr
import seaborn as sns
import ptitprince as pt
import pandas as pd
from plotly.subplots import make_subplots
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, plot
from IPython.core.display import display, HTML
Check maps available via neuromaps#
from neuromaps import datasets
datasets.available_annotations()
[('abagen', 'genepc1', 'fsaverage', '10k'),
('aghourian2017', 'feobv', 'MNI152', '1mm'),
('alarkurtti2015', 'raclopride', 'MNI152', '3mm'),
('bedard2019', 'feobv', 'MNI152', '1mm'),
('beliveau2017', 'az10419369', 'MNI152', '1mm'),
('beliveau2017', 'cimbi36', 'MNI152', '1mm'),
('beliveau2017', 'cumi101', 'MNI152', '1mm'),
('beliveau2017', 'dasb', 'MNI152', '1mm'),
('beliveau2017', 'sb207145', 'MNI152', '1mm'),
('ding2010', 'mrb', 'MNI152', '1mm'),
('dubois2015', 'abp688', 'MNI152', '1mm'),
('dukart2018', 'flumazenil', 'MNI152', '3mm'),
('dukart2018', 'fpcit', 'MNI152', '3mm'),
('fazio2016', 'madam', 'MNI152', '3mm'),
('finnema2016', 'ucbj', 'MNI152', '1mm'),
('gallezot2010', 'p943', 'MNI152', '1mm'),
('gallezot2017', 'gsk189254', 'MNI152', '1mm'),
('hcps1200', 'megalpha', 'fsLR', '4k'),
('hcps1200', 'megbeta', 'fsLR', '4k'),
('hcps1200', 'megdelta', 'fsLR', '4k'),
('hcps1200', 'meggamma1', 'fsLR', '4k'),
('hcps1200', 'meggamma2', 'fsLR', '4k'),
('hcps1200', 'megtheta', 'fsLR', '4k'),
('hcps1200', 'megtimescale', 'fsLR', '4k'),
('hcps1200', 'myelinmap', 'fsLR', '32k'),
('hcps1200', 'thickness', 'fsLR', '32k'),
('hesse2017', 'methylreboxetine', 'MNI152', '3mm'),
('hill2010', 'devexp', 'fsLR', '164k'),
('hill2010', 'evoexp', 'fsLR', '164k'),
('hillmer2016', 'flubatine', 'MNI152', '1mm'),
('jaworska2020', 'fallypride', 'MNI152', '1mm'),
('kaller2017', 'sch23390', 'MNI152', '3mm'),
('kantonen2020', 'carfentanil', 'MNI152', '3mm'),
('laurikainen2018', 'fmpepd2', 'MNI152', '1mm'),
('margulies2016', 'fcgradient01', 'fsLR', '32k'),
('margulies2016', 'fcgradient02', 'fsLR', '32k'),
('margulies2016', 'fcgradient03', 'fsLR', '32k'),
('margulies2016', 'fcgradient04', 'fsLR', '32k'),
('margulies2016', 'fcgradient05', 'fsLR', '32k'),
('margulies2016', 'fcgradient06', 'fsLR', '32k'),
('margulies2016', 'fcgradient07', 'fsLR', '32k'),
('margulies2016', 'fcgradient08', 'fsLR', '32k'),
('margulies2016', 'fcgradient09', 'fsLR', '32k'),
('margulies2016', 'fcgradient10', 'fsLR', '32k'),
('mueller2013', 'intersubjvar', 'fsLR', '164k'),
('naganawa2020', 'lsn3172176', 'MNI152', '1mm'),
('neurosynth', 'cogpc1', 'MNI152', '2mm'),
('norgaard2020', 'flumazenil', 'MNI152', '1mm'),
('normandin2015', 'omar', 'MNI152', '1mm'),
('radnakrishnan2018', 'gsk215083', 'MNI152', '1mm'),
('raichle', 'cbf', 'fsLR', '164k'),
('raichle', 'cbv', 'fsLR', '164k'),
('raichle', 'cmr02', 'fsLR', '164k'),
('raichle', 'cmruglu', 'fsLR', '164k'),
('reardon2018', 'scalinghcp', 'civet', '41k'),
('reardon2018', 'scalingnih', 'civet', '41k'),
('reardon2018', 'scalingpnc', 'civet', '41k'),
('rosaneto', 'abp688', 'MNI152', '1mm'),
('sandiego2015', 'flb457', 'MNI152', '1mm'),
('sasaki2012', 'fepe2i', 'MNI152', '1mm'),
('satterthwaite2014', 'meancbf', 'MNI152', '1mm'),
('savli2012', 'altanserin', 'MNI152', '3mm'),
('savli2012', 'dasb', 'MNI152', '3mm'),
('savli2012', 'p943', 'MNI152', '3mm'),
('savli2012', 'way100635', 'MNI152', '3mm'),
('smart2019', 'abp688', 'MNI152', '1mm'),
('smith2017', 'flb457', 'MNI152', '1mm'),
('sydnor2021', 'SAaxis', 'fsLR', '32k'),
('tuominen', 'feobv', 'MNI152', '2mm'),
('turtonen2020', 'carfentanil', 'MNI152', '1mm'),
('xu2020', 'FChomology', 'fsLR', '32k'),
('xu2020', 'evoexp', 'fsLR', '32k')]
maps to evaluate/use#
each map will be compared to the three gradients based on spatial null models (surrogate maps)
values of each map will be used to color the data points in the 3D scatterplot to evaluate if these maps/features therein follow task-based gradient (or vice-versa)
common space to utilize: fsaverage 10k
structure & genetics#
myelin -> hcp1200
thickness -> hcp1200
add MPC histology
PC1 of genes in Allen Human Brain Atlas -> abagen
metabolism#
cerebral blood flow -> raichle cbf
oxygen -> raichle cmr02
glucose -> raichle cmruglu
evolution & development#
evolutionary cortical expansion -> xu2020 evoexp
cross-species functional homology -> xu2020 fchomology
cortical areal scaling -> reardon2018 scalinghcp
electrophysiology#
alpha -> hcp1200
beta -> hcp1200
delta -> hcp1200
theta -> hcp1200
intrinsic timescale -> hcp1200
function#
rs-fmri intersubject variability -> mueller2013
gradient 1 -> margulies
sensory-association mean rank axis -> sydnor2021
get and check template#
fsaverage = datasets.fetch_fsaverage(density='10k')
fsaverage
{'white': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_white.surf.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_white.surf.gii')),
'pial': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_pial.surf.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_pial.surf.gii')),
'inflated': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_inflated.surf.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_inflated.surf.gii')),
'sphere': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_sphere.surf.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_sphere.surf.gii')),
'medial': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_desc-nomedialwall_dparc.label.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_desc-nomedialwall_dparc.label.gii')),
'sulc': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_desc-sulc_midthickness.shape.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_desc-sulc_midthickness.shape.gii')),
'vaavg': Surface(L=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_desc-vaavg_midthickness.shape.gii'), R=PosixPath('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_desc-vaavg_midthickness.shape.gii'))}
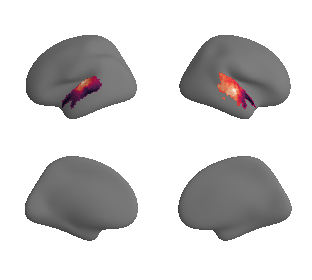
get and transform auditory cortex mask#
ac_mask = 'prerequisites/ac_rois/tpl-MNI152NLin6Sym_res_02_desc-AC_mask.nii.gz'
ac_mask_surface = transforms.mni152_to_fsaverage(ac_mask, '10k', method='nearest')
# check unique values
np.unique(ac_mask_surface[0].agg_data())
array([0., 1.], dtype=float32)
plot_surf_template((ac_mask_surface[0], ac_mask_surface[1]), 'fsaverage', '10k');

view_surf(fsaverage['inflated'][0], ac_mask_surface[0].agg_data(), threshold=0.5,
symmetric_cmap=False, cmap='Blues', colorbar=False)
view_surf(fsaverage['inflated'][1], ac_mask_surface[1].agg_data(), threshold=0.5,
symmetric_cmap=False, cmap='Blues', colorbar=False)
def mask_surface(surface_map, surface_mask):
# check if input is str, if so load gii
if isinstance(surface_map, str):
surface_map = nb.load(surface_map)
if isinstance(surface_mask, str):
surface_mask = nb.load(surface_mask)
# copy surface map data
surface_map_mask = deepcopy(surface_map.agg_data())
# mask surface map data based on provided mask
surface_map_mask[surface_mask.agg_data() == 0]=np.nan
# initiate new data array with masked surface map data
darray = nb.gifti.GiftiDataArray(surface_map_mask,
intent='NIFTI_INTENT_ESTIMATE',
datatype='NIFTI_TYPE_FLOAT32')
# create new gii image based on new darray
img = nb.GiftiImage(darrays=[darray])
# get surface data within mask
surface_data_masked = surface_map.agg_data()[np.where(surface_mask.agg_data() == 1)]
# return masked surface image & obtained surface data within mask
return img, surface_data_masked
get and transform neurosynth auditory cortex gradients#
gradient_1_surface = transforms.mni152_to_fsaverage('results/gradients/gradient-1_space-MNI152NLin6Sym_mask-ac.nii.gz', '10k', method='nearest')
gradient_2_surface = transforms.mni152_to_fsaverage('results/gradients/gradient-2_space-MNI152NLin6Sym_mask-ac.nii.gz', '10k', method='nearest')
gradient_3_surface = transforms.mni152_to_fsaverage('results/gradients/gradient-3_space-MNI152NLin6Sym_mask-ac.nii.gz', '10k', method='nearest')
gradient_1_map_left_fsaverage_mask_ac = np.empty_like(gradient_1_surface[0].agg_data())
gradient_1_map_left_fsaverage_mask_ac[ac_mask_surface[0].agg_data() == 0]=np.nan
gradient_1_map_right_fsaverage_mask_ac = np.empty_like(gradient_1_surface[1].agg_data())
gradient_1_map_right_fsaverage_mask_ac[ac_mask_surface[1].agg_data() == 0]=np.nan
gradient_1_map_left_fsaverage_mask_ac, gradient_1_map_left_fsaverage_mask_ac_data = mask_surface(gradient_1_surface[0], ac_mask_surface[0])
gradient_1_map_right_fsaverage_mask_ac, gradient_1_map_right_fsaverage_mask_ac_data = mask_surface(gradient_1_surface[1], ac_mask_surface[1])
plot_surf_template((gradient_1_map_left_fsaverage_mask_ac, gradient_1_map_right_fsaverage_mask_ac),
'fsaverage', '10k', cmap='crest');
structure & genetics#
myelin#
# get myelin maps
hcp_myelin_maps = datasets.fetch_annotation(source='hcps1200', desc='myelinmap')
# briefly check them
hcp_myelin_maps
['/Users/peerherholz/neuromaps-data/annotations/hcps1200/myelinmap/fsLR/source-hcps1200_desc-myelinmap_space-fsLR_den-32k_hemi-L_feature.func.gii',
'/Users/peerherholz/neuromaps-data/annotations/hcps1200/myelinmap/fsLR/source-hcps1200_desc-myelinmap_space-fsLR_den-32k_hemi-R_feature.func.gii']
# assign left and right map
hcp_myelin_map_left = hcp_myelin_maps[0]
hcp_myelin_map_right = hcp_myelin_maps[1]
# transform to fsaverage 10k
hcp_myelin_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_myelin_map_left, '10k', hemi='L')[0]
hcp_myelin_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_myelin_map_right, '10k', hemi='R')[0]
# check shape of map data
hcp_myelin_map_left_fsaverage.agg_data().shape
(10242,)
# plot whole-brain map
plot_surf_template((hcp_myelin_map_left_fsaverage, hcp_myelin_map_right_fsaverage), 'fsaverage', '10k', cmap='rocket');
view_surf(fsaverage['inflated'][0], hcp_myelin_map_left_fsaverage.agg_data(), cmap='rocket',
symmetric_cmap=False)
view_surf(fsaverage['inflated'][1], hcp_myelin_map_right_fsaverage.agg_data(), cmap='rocket',
symmetric_cmap=False)
# check number of myelin map vertices included in ac mask
hcp_myelin_map_left_fsaverage.agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)].shape
(434,)
# make sure it's the same number in the neurosynth ac gradient maps
gradient_1_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)].shape
(434,)
# mask whole-brain map with AC mask to get masked map and respective data - left hemisphere
hcp_myelin_map_left_fsaverage_mask_ac, hcp_myelin_map_left_fsaverage_mask_ac_data = mask_surface(hcp_myelin_map_left_fsaverage, ac_mask_surface[0])
# mask whole-brain map with AC mask to get masked map and respective data - right hemisphere
hcp_myelin_map_right_fsaverage_mask_ac, hcp_myelin_map_right_fsaverage_mask_ac_data = mask_surface(hcp_myelin_map_right_fsaverage, ac_mask_surface[1])
# plot masked map
plot_surf_template((hcp_myelin_map_left_fsaverage_mask_ac, hcp_myelin_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='rocket');
# correlate map with 1st gradient (within AC mask) - left hemisphere
grad_1_left_hcp_myelin_left_cor = stats.pearsonr(gradient_1_map_left_fsaverage_mask_ac_data,
hcp_myelin_map_left_fsaverage_mask_ac_data)
print('The maps are correlated with an r of %s at p = %s' %(str(grad_1_left_hcp_myelin_left_cor[0]),
str(grad_1_left_hcp_myelin_left_cor[1])))
The maps are correlated with an r of -0.7654862845200793 at p = 9.513620971538127e-85
# correlate map with 1st gradient (within AC mask) - right hemisphere
grad_1_right_hcp_myelin_right_cor = stats.pearsonr(gradient_1_map_right_fsaverage_mask_ac_data,
hcp_myelin_map_right_fsaverage_mask_ac_data)
print('The maps are correlated with an r of %s at p = %s' %(str(grad_1_right_hcp_myelin_right_cor[0]),
str(grad_1_right_hcp_myelin_right_cor[1])))
The maps are correlated with an r of -0.7569838780372673 at p = 2.257173511430903e-80
Short side quest: spatial null models ie surrogate maps#
# compute vertex distance matrix - left hemisphere
workbench.geo.cortex('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-L_inflated.surf.gii',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat.txt')
Running vertex 0 of 10242
Running vertex 1000 of 10242
Running vertex 2000 of 10242
Running vertex 3000 of 10242
Running vertex 4000 of 10242
Running vertex 5000 of 10242
Running vertex 6000 of 10242
Running vertex 7000 of 10242
Running vertex 8000 of 10242
Running vertex 9000 of 10242
Running vertex 10000 of 10242
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat.txt'
# compute vertex distance matrix - right hemisphere
workbench.geo.cortex('/Users/peerherholz/neuromaps-data/atlases/fsaverage/tpl-fsaverage_den-10k_hemi-R_inflated.surf.gii',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat.txt')
Running vertex 0 of 10242
Running vertex 1000 of 10242
Running vertex 2000 of 10242
Running vertex 3000 of 10242
Running vertex 4000 of 10242
Running vertex 5000 of 10242
Running vertex 6000 of 10242
Running vertex 7000 of 10242
Running vertex 8000 of 10242
Running vertex 9000 of 10242
Running vertex 10000 of 10242
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat.txt'
# load computed vertex distance matrix - left hemisphere
fsaverage_10k_hemi_L_desc_distmat = np.loadtxt('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat.txt')
# load computed vertex distance matrix - right hemisphere
fsaverage_10k_hemi_R_desc_distmat = np.loadtxt('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat.txt')
# check shape of computed vertex distance matrix - left hemisphere
fsaverage_10k_hemi_L_desc_distmat.shape
(10242, 10242)
# check shape of computed vertex distance matrix - left hemisphere
fsaverage_10k_hemi_R_desc_distmat.shape
(10242, 10242)
# restrict computed vertex distance matrix to vertices within AC mask - left hemisphere
fsaverage_10k_hemi_L_desc_distmat_AC = fsaverage_10k_hemi_L_desc_distmat[np.where(ac_mask_surface[0].agg_data() == 1),:].squeeze()[:, np.where(ac_mask_surface[0].agg_data() == 1)].squeeze()
# restrict computed vertex distance matrix to vertices within AC mask - left hemisphere
fsaverage_10k_hemi_R_desc_distmat_AC = fsaverage_10k_hemi_R_desc_distmat[np.where(ac_mask_surface[1].agg_data() == 1),:].squeeze()[:, np.where(ac_mask_surface[1].agg_data() == 1)].squeeze()
# check shape of computed vertex distance matrix within AC mask - left hemisphere
fsaverage_10k_hemi_L_desc_distmat_AC.shape
(434, 434)
# check shape of computed vertex distance matrix within AC mask - right hemisphere
fsaverage_10k_hemi_R_desc_distmat_AC.shape
(426, 426)
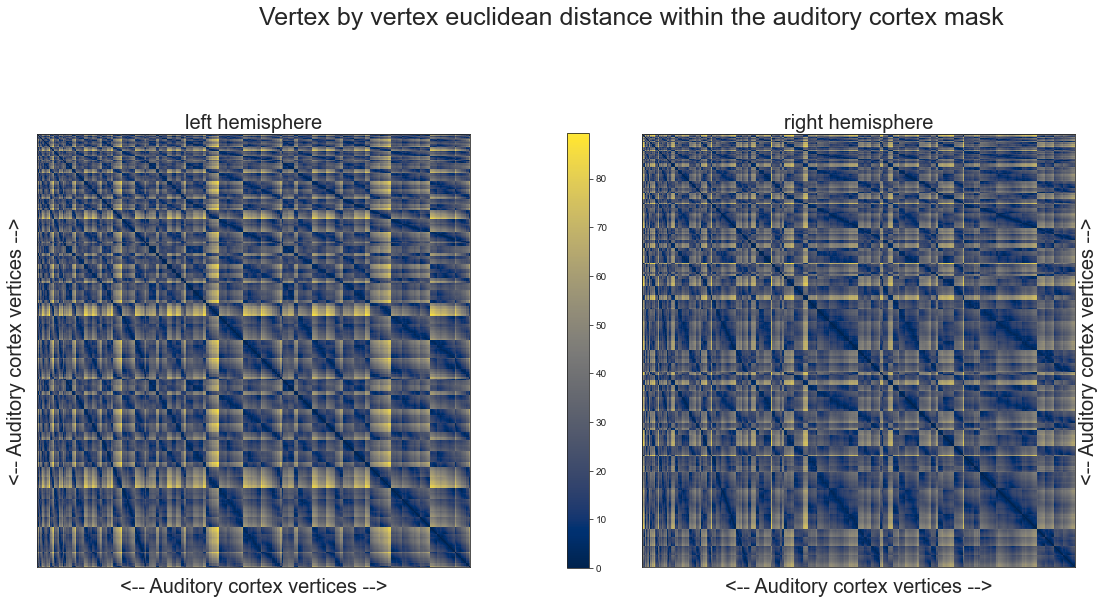
def plot_distance_matrix(dist_mat_left, dist_mat_right, interactive=False):
if interactive is False:
sns.set_style('white')
plt.rcParams["font.family"] = "Arial"
fig, axes = plt.subplots(1,2, figsize = (22, 10))
plot_matrix(dist_mat_left, cmap='cividis', axes=axes[0])
axes[0].set_xlabel('<-- Auditory cortex vertices -->', fontsize=20)
axes[0].set_xticklabels([''])
axes[0].set_ylabel('<-- Auditory cortex vertices -->', fontsize=20)
axes[0].set_yticklabels([''])
axes[0].set_title('left hemisphere', fontsize=20)
plt.subplots_adjust(wspace=0.4)
plot_matrix(dist_mat_right, cmap='cividis', axes=axes[1], colorbar=False)
axes[1].set_xlabel('<-- Auditory cortex vertices -->', fontsize=20)
axes[1].set_xticklabels([''])
axes[1].yaxis.set_label_position("right")
axes[1].set_ylabel('<-- Auditory cortex vertices -->', fontsize=20)
axes[1].set_yticklabels([''])
axes[1].set_title('right hemisphere', fontsize=20)
fig.suptitle('Vertex by vertex euclidean distance within the auditory cortex mask', size=25)
plt.show()
else:
fig = make_subplots(rows=1, cols=2,
subplot_titles=('left hemisphere', 'right hemisphere'),
horizontal_spacing=0.18)
fig.add_trace(go.Heatmap(z=dist_mat_left[:630,:630],
colorscale='cividis',colorbar_x=0.46, transpose=True), row=1, col=1)
fig.add_trace(go.Heatmap(z=dist_mat_right[:630,:630],
colorscale='cividis', showscale = False, transpose=True), row=1, col=2)
fig.update_layout(
autosize=False,
width=900,
height=600,)
fig['layout'].update(template='simple_white')
fig.update_layout(
title={
'text': "Vertex by Vertex euclidean distance within the auditory cortex mask",
'y':0.93,
'x':0.5,
'xanchor': 'center',
'yanchor': 'top'})
fig['layout']['xaxis']['title']='<-- Auditory cortex vertices -->'
fig['layout']['xaxis2']['title']='<-- Auditory cortex vertices -->'
fig['layout']['yaxis']['title']='<-- Auditory cortex vertices -->'
fig['layout']['yaxis2']['title']='<-- Auditory cortex vertices -->'
fig['layout']['yaxis']['autorange'] = "reversed"
fig['layout']['yaxis2']['autorange'] = "reversed"
fig['layout']['yaxis2']['side'] = "right"
init_notebook_mode(connected=True)
plot(fig, filename = 'vertex2vertex_AC_dist.html')
display(HTML('vertex2vertex_AC_dist.html'))
plot_distance_matrix(fsaverage_10k_hemi_L_desc_distmat_AC, fsaverage_10k_hemi_R_desc_distmat_AC)
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/ipykernel_launcher.py:15: UserWarning:
FixedFormatter should only be used together with FixedLocator
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/ipykernel_launcher.py:17: UserWarning:
FixedFormatter should only be used together with FixedLocator
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/ipykernel_launcher.py:24: UserWarning:
FixedFormatter should only be used together with FixedLocator
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/ipykernel_launcher.py:27: UserWarning:
FixedFormatter should only be used together with FixedLocator
# save vertex distance matrix within AC mask
np.savetxt('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask.txt',
fsaverage_10k_hemi_L_desc_distmat_AC)
np.savetxt('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask.txt',
fsaverage_10k_hemi_R_desc_distmat_AC)
# created a memory-mapped object to use for surrogate maps based on dense data - left hemisphere
fsaverage_10k_hemi_L_desc_distmat_AC_mapped = txt2memmap('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask.txt',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/', maskfile=None, delimiter=' ')
fsaverage_10k_hemi_L_desc_distmat_AC_mapped
{'distmat': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/distmat.npy',
'index': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/index.npy'}
os.rename('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/distmat.npy',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask.npy')
os.rename('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/index.npy',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask_index.npy')
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['distmat'] = '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask.npy'
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['index'] = '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask_index.npy'
fsaverage_10k_hemi_L_desc_distmat_AC_mapped
{'distmat': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask.npy',
'index': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-L_inflated_desc-distmat_AC_mask_index.npy'}
# created a memory-mapped object to use for surrogate maps based on dense data - right hemisphere
fsaverage_10k_hemi_R_desc_distmat_AC_mapped = txt2memmap('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask.txt',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/', maskfile=None, delimiter=' ')
os.rename('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/distmat.npy',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask.npy')
os.rename('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/index.npy',
'/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask_index.npy')
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['distmat'] = '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask.npy'
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['index'] = '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask_index.npy'
fsaverage_10k_hemi_R_desc_distmat_AC_mapped
{'distmat': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask.npy',
'index': '/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/tpl-fsaverage_den-10k_hemi-R_inflated_desc-distmat_AC_mask_index.npy'}
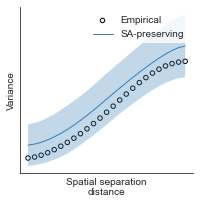
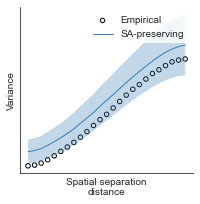
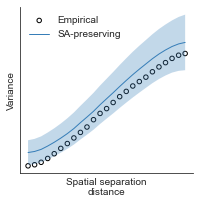
# evaluate fit of surrogate maps re spatial autocorrelation via variograms - left hemisphere
sampled_fit(hcp_myelin_map_left_fsaverage_mask_ac_data,
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['index'],
ns=50, knn=400, seed=42, kernel='gaussian', nsurr=1000)
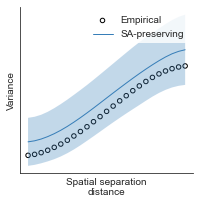
# evaluate fit of surrogate maps re spatial autocorrelation via variograms - right hemisphere
sampled_fit(hcp_myelin_map_right_fsaverage_mask_ac_data,
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['index'],
ns=50, knn=400, seed=42, kernel='gaussian', nsurr=1000)
def compute_variograms(surface_map_data_lh, surface_map_data_rh, ns=50, knn=400):
print('Computing variogram for left hemisphere')
sampled_fit(surface_map_data_lh,
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['index'],
ns=ns, knn=knn, seed=42, kernel='gaussian', nsurr=1000)
print('Computing variogram for right hemisphere')
sampled_fit(surface_map_data_rh,
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['index'],
ns=ns, knn=knn, seed=42, kernel='gaussian', nsurr=1000)
# set up Sampled object to create surrogate maps - left hemisphere
sampled_AC_left = Sampled(hcp_myelin_map_left_fsaverage_mask_ac_data,
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['index'],
ns=50, knn=400, seed=42,
kernel='gaussian')
# create 1k surrogate maps - left hemisphere
hcp_myelin_map_left_fsaverage_mask_ac_surrogates = sampled_AC_left(n=1000)
# set up Sampled object to create surrogate maps - right hemisphere
sampled_AC_right = Sampled(hcp_myelin_map_right_fsaverage_mask_ac_data,
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['index'],
ns=50, knn=400, seed=42,
kernel='gaussian')
# create 1k surrogate maps - right hemisphere
hcp_myelin_map_right_fsaverage_mask_ac_surrogates = sampled_AC_right(n=1000)
def compute_surrogate_maps(surface_map_data_lh, surface_map_data_rh, ns=50, knn=400):
sampled_AC_left = Sampled(surface_map_data_lh,
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_L_desc_distmat_AC_mapped['index'],
ns=ns, knn=knn, seed=42, kernel='gaussian')
sampled_AC_right = Sampled(surface_map_data_rh,
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['distmat'],
fsaverage_10k_hemi_R_desc_distmat_AC_mapped['index'],
ns=ns, knn=knn, seed=42, kernel='gaussian')
print('working on LH surrogates')
sampled_AC_left_surrogates = sampled_AC_left(n=1000)
print('working on RH surrogates')
sampled_AC_right_surrogates = sampled_AC_right(n=1000)
return sampled_AC_left_surrogates, sampled_AC_right_surrogates
# plot first ten surrogate maps
for sur in range(0,10):
sur_tmp_left = deepcopy(ac_mask_surface[0].agg_data())
sur_tmp_left[ac_mask_surface[0].agg_data() == 1] = hcp_myelin_map_left_fsaverage_mask_ac_surrogates[sur]
# initiate new data array with masked surface map data
darray_left = nb.gifti.GiftiDataArray(sur_tmp_left,
intent='NIFTI_INTENT_ESTIMATE',
datatype='NIFTI_TYPE_FLOAT32')
# create new gii image based on new darray
sur_tmp_left = nb.GiftiImage(darrays=[darray_left])
sur_tmp_right = deepcopy(ac_mask_surface[1].agg_data())
sur_tmp_right[ac_mask_surface[1].agg_data() == 1] = hcp_myelin_map_right_fsaverage_mask_ac_surrogates[sur]
# initiate new data array with masked surface map data
darray_right = nb.gifti.GiftiDataArray(sur_tmp_right,
intent='NIFTI_INTENT_ESTIMATE',
datatype='NIFTI_TYPE_FLOAT32')
# create new gii image based on new darray
sur_tmp_right = nb.GiftiImage(darrays=[darray_right])
plot_surf_template((sur_tmp_left, sur_tmp_right), 'fsaverage', '10k', cmap='rocket');

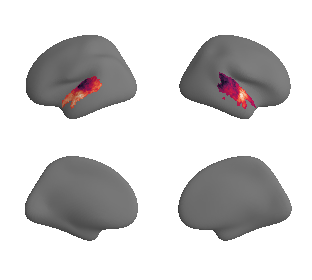
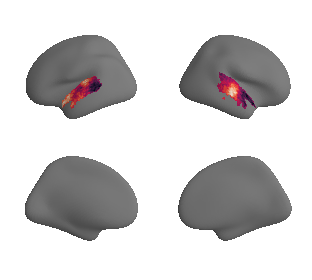
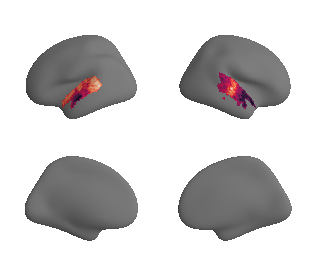
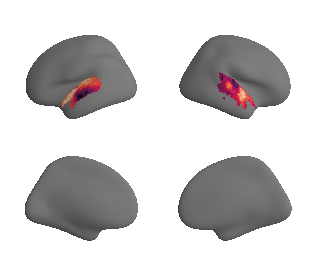
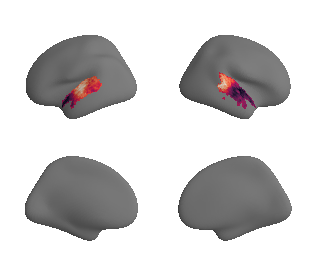
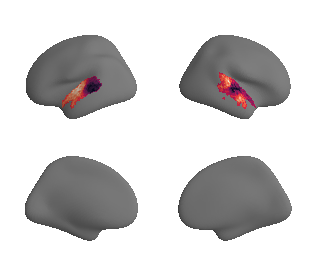
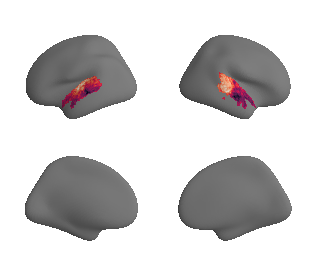
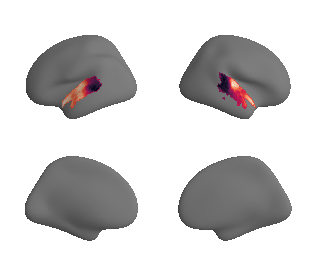
def plot_surrogate_maps(sampled_AC_left_surrogates, sampled_AC_right_surrogates, n_sur=10, cmap='rocket'):
for sur in range(0,n_sur):
sur_tmp_left = deepcopy(ac_mask_surface[0].agg_data())
sur_tmp_left[ac_mask_surface[0].agg_data() == 1] = sampled_AC_left_surrogates[sur]
# initiate new data array with masked surface map data
darray_left = nb.gifti.GiftiDataArray(sur_tmp_left,
intent='NIFTI_INTENT_ESTIMATE',
datatype='NIFTI_TYPE_FLOAT32')
# create new gii image based on new darray
sur_tmp_left = nb.GiftiImage(darrays=[darray_left])
sur_tmp_right = deepcopy(ac_mask_surface[1].agg_data())
sur_tmp_right[ac_mask_surface[1].agg_data() == 1] = sampled_AC_right_surrogates[sur]
# initiate new data array with masked surface map data
darray_right = nb.gifti.GiftiDataArray(sur_tmp_right,
intent='NIFTI_INTENT_ESTIMATE',
datatype='NIFTI_TYPE_FLOAT32')
# create new gii image based on new darray
sur_tmp_right = nb.GiftiImage(darrays=[darray_right])
plot_surf_template((sur_tmp_left, sur_tmp_right), 'fsaverage', '10k', cmap=cmap);
# compute correlation between 1st gradient and surrogate maps - left hemisphere
surrogate_brainmap_corrs_left = pearsonr(gradient_1_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)],
hcp_myelin_map_left_fsaverage_mask_ac_surrogates).flatten()
# compute correlation between 1st gradient and surrogate maps - right hemisphere
surrogate_brainmap_corrs_right = pearsonr(gradient_1_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)],
hcp_myelin_map_right_fsaverage_mask_ac_surrogates).flatten()
# compare correlation between correlation of non-permuted maps to distribution based on surrogate maps to get p value
grad_1_left_hcp_myelin_left_cor = stats.pearsonr(gradient_1_map_left_fsaverage_mask_ac_data,
hcp_myelin_map_left_fsaverage_mask_ac_data)[0]
grad_1_left_hcp_myelin_left_cor_p = nonparp(grad_1_left_hcp_myelin_left_cor,
surrogate_brainmap_corrs_left)
print('The LH maps are correlated with an r of %s at p = %s' %(str(grad_1_left_hcp_myelin_left_cor),
str(grad_1_left_hcp_myelin_left_cor_p)))
grad_1_right_hcp_myelin_right_cor = stats.pearsonr(gradient_1_map_right_fsaverage_mask_ac_data,
hcp_myelin_map_right_fsaverage_mask_ac_data)[0]
grad_1_right_hcp_myelin_right_cor_p = nonparp(grad_1_right_hcp_myelin_right_cor,
surrogate_brainmap_corrs_right)
print('The RH maps are correlated with an r of %s at p = %s' %(str(grad_1_right_hcp_myelin_right_cor),
str(grad_1_right_hcp_myelin_right_cor_p)))
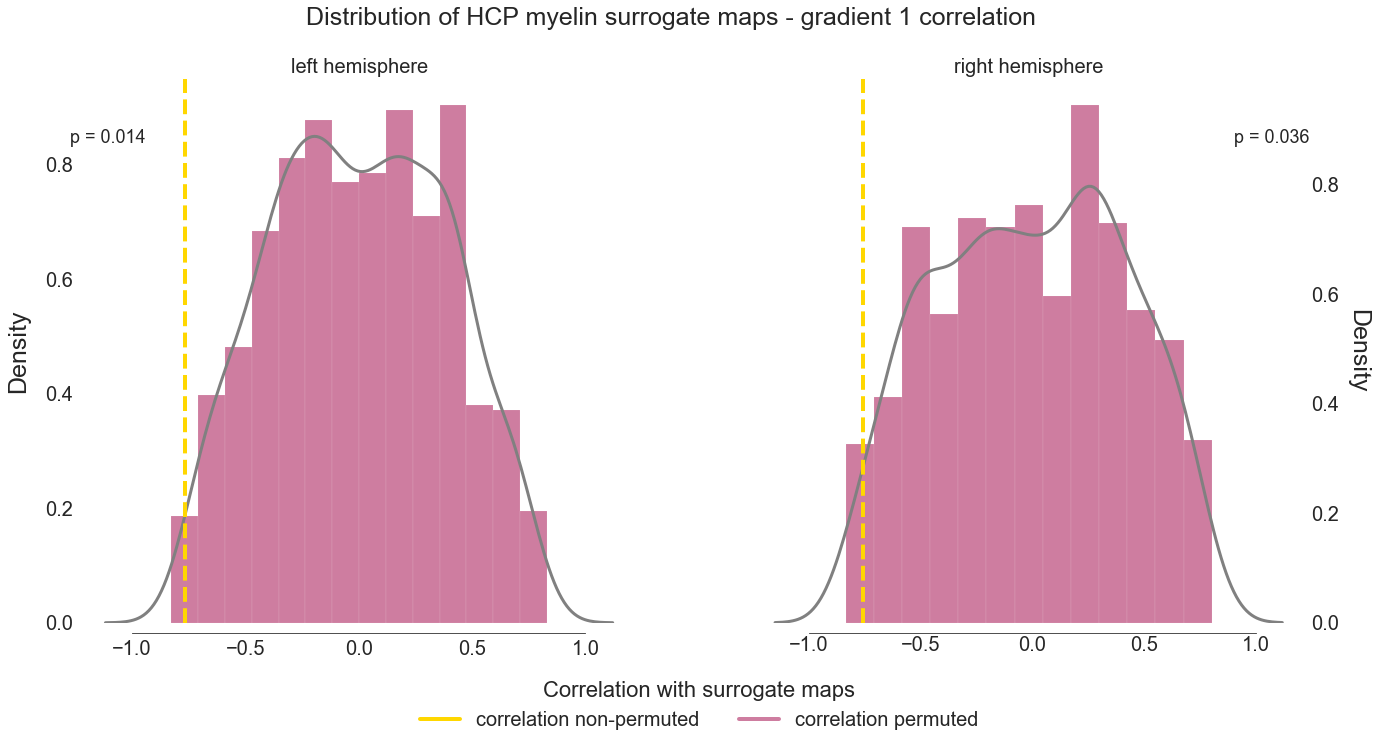
The LH maps are correlated with an r of -0.7654862845200793 at p = 0.014
The RH maps are correlated with an r of -0.7569838780372673 at p = 0.036
# plot correlation based on non-permuted maps and distribution based on surrogate maps
sns.set_style('white')
plt.rcParams["font.family"] = "Arial"
fig, axes = plt.subplots(1,2, figsize = (22, 10))
sns.distplot(surrogate_brainmap_corrs_left, ax=axes[0],
kde_kws={"color": "grey", "lw": 3},
hist_kws={"linewidth": 0.1,
"alpha": 1, "color": "#CE7Da0"})
#axes[0].set_xlabel('Correlation with surrogate maps', fontsize=25)
axes[0].tick_params(labelsize=20)
axes[0].set_ylabel('Density', fontsize=25, labelpad=15)
axes[0].set_title('left hemisphere', fontsize=20)
axes[0].axvline(grad_1_left_hcp_myelin_left_cor, 0, 1.8, color='gold', linestyle='dashed', lw=4)
plt.text(-4.3, 0.875, 'p = %s' %round(nonparp(grad_1_left_hcp_myelin_left_cor, surrogate_brainmap_corrs_left), 3),
fontsize=18)
plt.subplots_adjust(wspace=0.2)
sns.distplot(surrogate_brainmap_corrs_right, ax=axes[1],
kde_kws={"color": "grey", "lw": 3},
hist_kws={"linewidth": 0.1,
"alpha": 1, "color": "#CE7Da0"})
#axes[1].set_xlabel('Correlation with surrogate maps', fontsize=25)
axes[1].tick_params(labelsize=20)
axes[1].set_ylabel('Density', fontsize=25, rotation=270, labelpad=30)
axes[1].set_title('right hemisphere', fontsize=20)
axes[1].tick_params(axis=u'both', which=u'both',length=0)
axes[1].yaxis.tick_right()
axes[1].yaxis.set_label_position("right")
axes[1].axvline(grad_1_right_hcp_myelin_right_cor, 0, 1.8, color='gold', linestyle='dashed', lw=4)
plt.text(0.9, 0.875, 'p = %s' %round(nonparp(grad_1_right_hcp_myelin_right_cor, surrogate_brainmap_corrs_right), 3),
fontsize=18)
fig.suptitle('Distribution of HCP myelin surrogate maps - gradient 1 correlation ', size=25)
legend = [Line2D([0], [0], color='gold', lw=4, label='correlation non-permuted'),
Line2D([0], [0], color="#CE7Da0", lw=4, label='correlation permuted')]
plt.legend(handles=legend, bbox_to_anchor=(-0.09, -0.23), fontsize=20,
ncol=2, loc="lower center", frameon=False, title='Correlation with surrogate maps',
title_fontsize=22)
sns.despine(left=True, offset=10, trim=True)
plt.show()
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/seaborn/distributions.py:2551: FutureWarning:
`distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
/Users/peerherholz/anaconda3/envs/ns_ac_walkthrough/lib/python3.7/site-packages/seaborn/distributions.py:2551: FutureWarning:
`distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
# compute correlation between all gradients and surrogate maps
grad_hcp_myelin_cors = {}
for i, hemi in enumerate(['LH', 'RH']):
print('Working on %s maps \n' %hemi)
grad_hcp_myelin_cors[hemi] = {}
for i_grad,grad in enumerate([gradient_1_surface, gradient_2_surface, gradient_3_surface]):
print('Working on gradient %s \n' %str(i_grad+1))
gradient_tmp_fsaverage_mask_ac, gradient_tmp_fsaverage_mask_ac_data = mask_surface(grad[i],
ac_mask_surface[i])
if hemi=='LH':
surrogate_brainmap_corrs = pearsonr(gradient_tmp_fsaverage_mask_ac_data,
hcp_myelin_map_left_fsaverage_mask_ac_surrogates).flatten()
surrogate_brainmap_corrs_r = stats.pearsonr(gradient_tmp_fsaverage_mask_ac_data,
hcp_myelin_map_left_fsaverage_mask_ac_data)[0]
elif hemi=='RH':
surrogate_brainmap_corrs = pearsonr(gradient_tmp_fsaverage_mask_ac_data,
hcp_myelin_map_right_fsaverage_mask_ac_surrogates).flatten()
surrogate_brainmap_corrs_r = stats.pearsonr(gradient_tmp_fsaverage_mask_ac_data,
hcp_myelin_map_right_fsaverage_mask_ac_data)[0]
surrogate_brainmap_corrs_p = nonparp(surrogate_brainmap_corrs_r,
surrogate_brainmap_corrs)
print('The %s maps of gradient %s & HCP myelin are correlated with an r of %s at p = %s \n' %(hemi,
str(i_grad+1), str(surrogate_brainmap_corrs_r),
str(surrogate_brainmap_corrs_p)))
grad_hcp_myelin_cors[hemi]['grad_' + str(i_grad+1)] = {'r': surrogate_brainmap_corrs_r,
'p': surrogate_brainmap_corrs_p,
'cors': list(surrogate_brainmap_corrs)}
Working on LH maps
Working on gradient 1
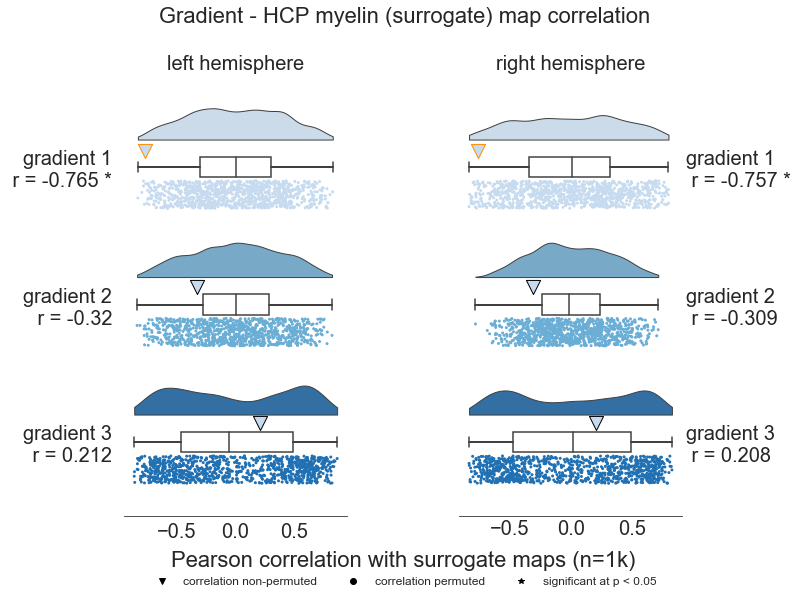
The LH maps of gradient 1 & HCP myelin are correlated with an r of -0.7654862845200793 at p = 0.014
Working on gradient 2
The LH maps of gradient 2 & HCP myelin are correlated with an r of -0.32037378678875467 at p = 0.449
Working on gradient 3
The LH maps of gradient 3 & HCP myelin are correlated with an r of 0.2117213791593506 at p = 0.817
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & HCP myelin are correlated with an r of -0.7569838780372673 at p = 0.036
Working on gradient 2
The RH maps of gradient 2 & HCP myelin are correlated with an r of -0.3092368007374292 at p = 0.367
Working on gradient 3
The RH maps of gradient 3 & HCP myelin are correlated with an r of 0.2081869879427032 at p = 0.82
# save correlation dictionary
json.dump(grad_hcp_myelin_cors, open('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_hcp_myelin_cors.json', 'w' ))
def compare_brain_maps_surrogates(surface_map_data_lh, surface_map_data_rh,
surface_map_data_lh_surrogates, surface_map_data_rh_surrogates,
feature='feature_name', path=os.curdir):
grad_feature_cors = {}
for i, hemi in enumerate(['LH', 'RH']):
print('Working on %s maps \n' %hemi)
grad_feature_cors[hemi] = {}
for i_grad,grad in enumerate([gradient_1_surface, gradient_2_surface, gradient_3_surface]):
print('Working on gradient %s \n' %str(i_grad+1))
gradient_tmp_fsaverage_mask_ac, gradient_tmp_fsaverage_mask_ac_data = mask_surface(grad[i],
ac_mask_surface[i])
if hemi=='LH':
surrogate_brainmap_corrs = pearsonr(gradient_tmp_fsaverage_mask_ac_data,
surface_map_data_lh_surrogates).flatten()
surrogate_brainmap_corrs_r = stats.pearsonr(gradient_tmp_fsaverage_mask_ac_data,
surface_map_data_lh)[0]
elif hemi=='RH':
surrogate_brainmap_corrs = pearsonr(gradient_tmp_fsaverage_mask_ac_data,
surface_map_data_rh_surrogates).flatten()
surrogate_brainmap_corrs_r = stats.pearsonr(gradient_tmp_fsaverage_mask_ac_data,
surface_map_data_rh)[0]
surrogate_brainmap_corrs_p = nonparp(surrogate_brainmap_corrs_r,
surrogate_brainmap_corrs)
print('The %s maps of gradient %s & %s are correlated with an r of %s at p = %s \n' %(hemi,
str(i_grad+1), feature, str(surrogate_brainmap_corrs_r),
str(surrogate_brainmap_corrs_p)))
grad_feature_cors[hemi]['grad_' + str(i_grad+1)] = {'r': surrogate_brainmap_corrs_r,
'p': surrogate_brainmap_corrs_p,
'cors': list(surrogate_brainmap_corrs)}
print('The results will be save to %s' %path)
json.dump(grad_feature_cors, open(path, 'w' ))
return grad_feature_cors
grad_hcp_myelin_cors = json.load(open('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_hcp_myelin_cors.json'))
# transform correlation dictionary to a pd.Dataframe - left hemisphere
grad_hcp_myelin_cors_df_left = pd.DataFrame({'grad': ['gradient 1'] * len(grad_hcp_myelin_cors['LH']['grad_1']['cors']) + \
['gradient 2'] * len(grad_hcp_myelin_cors['LH']['grad_2']['cors']) + \
['gradient 3'] * len(grad_hcp_myelin_cors['LH']['grad_3']['cors']),
'cors': grad_hcp_myelin_cors['LH']['grad_1']['cors'] + \
grad_hcp_myelin_cors['LH']['grad_2']['cors'] + \
grad_hcp_myelin_cors['LH']['grad_3']['cors']})
# transform correlation dictionary to a pd.Dataframe - right hemisphere
grad_hcp_myelin_cors_df_right = pd.DataFrame({'grad': ['gradient 1'] * len(grad_hcp_myelin_cors['RH']['grad_1']['cors']) + \
['gradient 2'] * len(grad_hcp_myelin_cors['RH']['grad_2']['cors']) + \
['gradient 3'] * len(grad_hcp_myelin_cors['RH']['grad_3']['cors']),
'cors': grad_hcp_myelin_cors['RH']['grad_1']['cors'] + \
grad_hcp_myelin_cors['RH']['grad_2']['cors'] + \
grad_hcp_myelin_cors['RH']['grad_3']['cors']})
# plot correlation based on non-permuted maps and distribution based on surrogate maps for all gradients
dx="grad"; dy="cors"; ort="h"; pal = "Set2"; sigma = .2
f, ax = plt.subplots(1,2, figsize=(10, 8))
ax[0]=pt.RainCloud(x = dx, y = dy, data = grad_hcp_myelin_cors_df_left, palette = 'Blues', bw = sigma,
width_viol = .5, ax = ax[0], orient = ort, move = .2)#, **font)
y = -0.12
for grad in ['grad_1', 'grad_2', 'grad_3']:
if grad_hcp_myelin_cors['LH'][grad]['p'] < 0.05:
ec = 'darkorange'
else:
ec = 'black'
ax[0].scatter(grad_hcp_myelin_cors['LH'][grad]['r'], y, marker='v', color=sns.color_palette('Blues', 3)[0],
edgecolor=ec, s=200)#, label='gradient 1: %.3f' %grad_hcp_myelin_cors['LH']['grad_1']['r'])
y = y + 0.99
ax[0].tick_params(labelsize=20)
ax[0].set_ylabel('', fontsize=25, labelpad=15)
ax[0].set_title('left hemisphere', fontsize=20)
ax[0].set_xlabel('')
list_yticklabels_left = ['gradient %s\n r = %s *'%(grad, str(round(grad_hcp_myelin_cors['LH']['grad_'+grad]['r'], 3))) if grad_hcp_myelin_cors['LH']['grad_'+grad]['p'] < 0.05 else 'gradient %s\n r = %s'%(grad, str(round(grad_hcp_myelin_cors['LH']['grad_'+grad]['r'], 3))) for grad in ['1', '2', '3']]
ax[0].set_yticklabels(list_yticklabels_left)
sns.despine(offset=5)
plt.subplots_adjust(wspace=0.5)
ax[1]=pt.RainCloud(x = dx, y = dy, data = grad_hcp_myelin_cors_df_right, palette = 'Blues', bw = sigma,
width_viol = .5, ax = ax[1], orient = ort, move = .2)#, **font)
y = -0.12
for grad in ['grad_1', 'grad_2', 'grad_3']:
if grad_hcp_myelin_cors['RH'][grad]['p'] < 0.05:
ec = 'darkorange'
else:
ec = 'black'
ax[1].scatter(grad_hcp_myelin_cors['RH'][grad]['r'], y, marker='v', color=sns.color_palette('Blues', 3)[0],
edgecolor=ec, s=200)#, label='gradient 1: %.3f' %grad_hcp_myelin_cors['LH']['grad_1']['r'])
y = y + 0.99
ax[1].tick_params(labelsize=20)
ax[1].set_ylabel('', fontsize=25, labelpad=15)
ax[1].set_title('right hemisphere', fontsize=20)
ax[1].set_xlabel('')
ax[1].tick_params(axis=u'both', which=u'both',length=0)
ax[1].yaxis.tick_right()
ax[1].yaxis.set_label_position("right")
list_yticklabels_right = ['gradient %s\n r = %s *'%(grad, str(round(grad_hcp_myelin_cors['RH']['grad_'+grad]['r'], 3))) if grad_hcp_myelin_cors['RH']['grad_'+grad]['p'] < 0.05 else 'gradient %s\n r = %s'%(grad, str(round(grad_hcp_myelin_cors['RH']['grad_'+grad]['r'], 3))) for grad in ['1', '2', '3']]
ax[1].set_yticklabels(list_yticklabels_right)
sns.despine(offset=5, left=True)
plt.legend()
plt.suptitle('Gradient - HCP myelin (surrogate) map correlation', fontsize=22, y=1, x=0.515)
legend = [Line2D([], [], color='black', marker='v', linestyle='None', label='correlation non-permuted'),
Line2D([], [], color='black', marker='o', linestyle='None', label='correlation permuted'),
Line2D([], [], color='black', marker='*', linestyle='None', label='significant at p < 0.05')]
plt.legend(handles=legend, bbox_to_anchor=(-0.25, -0.20), fontsize=12,
ncol=3, loc="lower center", frameon=False, title='Pearson correlation with surrogate maps (n=1k)',
title_fontsize=22)
No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument.
<matplotlib.legend.Legend at 0x7fc8806cdac8>
def plot_brain_surrogate_map_cor(grad_feature_cors, feature='feature_name'):
if isinstance(grad_feature_cors, str):
grad_feature_cors = json.load(open(grad_feature_cors))
grad_feature_cors_df_left = pd.DataFrame({'grad': ['gradient 1'] * len(grad_feature_cors['LH']['grad_1']['cors']) + \
['gradient 2'] * len(grad_feature_cors['LH']['grad_2']['cors']) + \
['gradient 3'] * len(grad_feature_cors['LH']['grad_3']['cors']),
'cors': grad_feature_cors['LH']['grad_1']['cors'] + \
grad_feature_cors['LH']['grad_2']['cors'] + \
grad_feature_cors['LH']['grad_3']['cors']})
grad_feature_cors_df_right = pd.DataFrame({'grad': ['gradient 1'] * len(grad_feature_cors['RH']['grad_1']['cors']) + \
['gradient 2'] * len(grad_feature_cors['RH']['grad_2']['cors']) + \
['gradient 3'] * len(grad_feature_cors['RH']['grad_3']['cors']),
'cors': grad_feature_cors['RH']['grad_1']['cors'] + \
grad_feature_cors['RH']['grad_2']['cors'] + \
grad_feature_cors['RH']['grad_3']['cors']})
dx="grad"; dy="cors"; ort="h"; pal = "Set2"; sigma = .2
f, ax = plt.subplots(1,2, figsize=(10, 8))
ax[0]=pt.RainCloud(x = dx, y = dy, data = grad_feature_cors_df_left, palette = 'Blues', bw = sigma,
width_viol = .5, ax = ax[0], orient = ort, move = .2)#, **font)
y = -0.12
for grad in ['grad_1', 'grad_2', 'grad_3']:
if grad_feature_cors_df_left['LH'][grad]['p'] < 0.05:
ec = 'darkorange'
else:
ec = 'black'
ax[0].scatter(grad_feature_cors_df_left['LH'][grad]['r'], y, marker='v', color=sns.color_palette('Blues', 3)[0],
edgecolor=ec, s=200)#, label='gradient 1: %.3f' %grad_hcp_myelin_cors['LH']['grad_1']['r'])
y = y + 0.99
ax[0].tick_params(labelsize=20)
ax[0].set_ylabel('', fontsize=25, labelpad=15)
ax[0].set_title('left hemisphere', fontsize=20)
ax[0].set_xlabel('')
ax[0].set_yticklabels(['gradient %s\n r = %s'%(grad, str(round(grad_feature_cors_df_left['LH']['grad_'+grad]['r'], 3))) for grad in ['1', '2', '3']])
sns.despine(offset=5)
plt.subplots_adjust(wspace=0.5)
ax[1]=pt.RainCloud(x = dx, y = dy, data = grad_feature_cors_df_right, palette = 'Blues', bw = sigma,
width_viol = .5, ax = ax[1], orient = ort, move = .2)#, **font)
y = -0.12
for grad in ['grad_1', 'grad_2', 'grad_3']:
if grad_feature_cors_df_right['RH'][grad]['p'] < 0.05:
ec = 'darkorange'
else:
ec = 'black'
ax[1].scatter(grad_feature_cors_df_right['RH'][grad]['r'], y, marker='v', color=sns.color_palette('Blues', 3)[0],
edgecolor=ec, s=200)#, label='gradient 1: %.3f' %grad_hcp_myelin_cors['LH']['grad_1']['r'])
y = y + 0.99
ax[1].tick_params(labelsize=20)
ax[1].set_ylabel('', fontsize=25, labelpad=15)
ax[1].set_title('right hemisphere', fontsize=20)
ax[1].set_xlabel('')
ax[1].tick_params(axis=u'both', which=u'both',length=0)
ax[1].yaxis.tick_right()
ax[1].yaxis.set_label_position("right")
ax[1].set_yticklabels(['gradient %s\n r = %s'%(grad, str(round(grad_feature_cors_df_right['RH']['grad_'+grad]['r'], 3))) for grad in ['1', '2', '3']])
sns.despine(offset=5, left=True)
plt.legend()
plt.suptitle('Gradient - (surrogate) %s map correlation' %feature, fontsize=22, y=1, x=0.515)
legend = [Line2D([], [], color='black', marker='v', linestyle='None', label='correlation non-permuted'),
Line2D([], [], color='black', marker='o', linestyle='None', label='correlation permuted'),
Line2D([0], [0], color='darkorange', lw=2, label='significant at p < 0.05'),]
plt.legend(handles=legend, bbox_to_anchor=(-0.25, -0.20), fontsize=12,
ncol=3, loc="lower center", frameon=False, title='Pearson correlation with surrogate maps (n=1k)',
title_fontsize=22)
return grad_feature_cors_df_left, grad_feature_cors_df_right
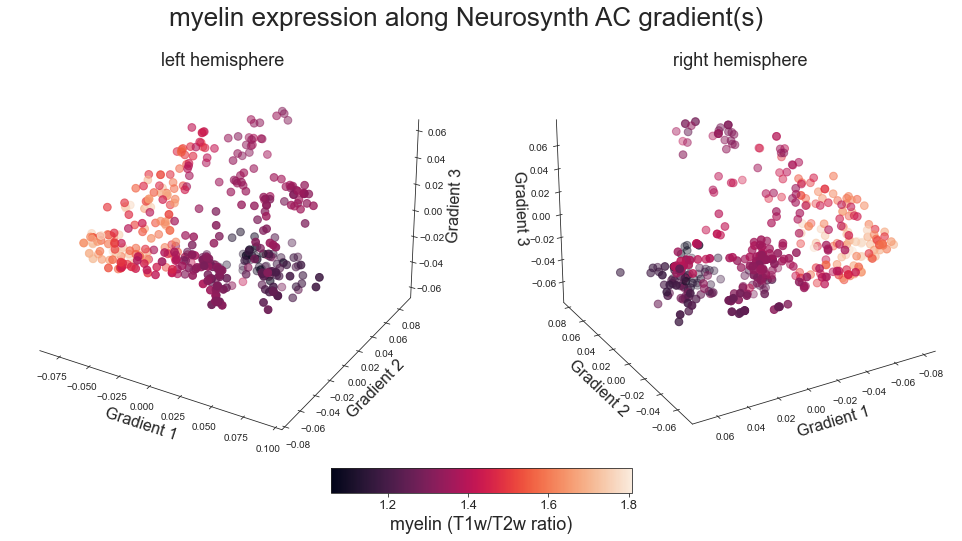
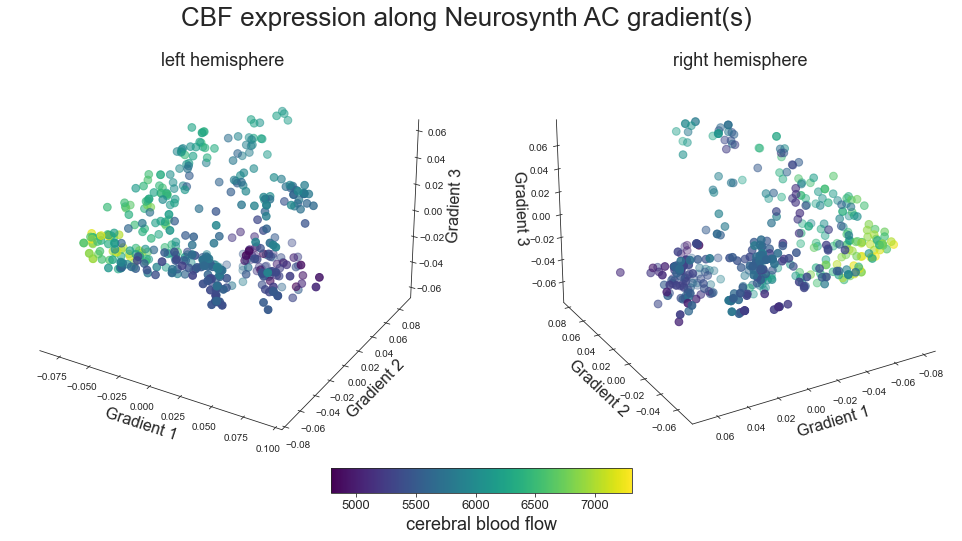
# plot map data (ie feature expression per vertex) in space spanned by the gradients - left hemisphere
sns.set_style('white')
plt.rcParams["font.family"] = "Arial"
fig = plt.figure(figsize=(13,10))
ax = plt.axes(projection="3d")
ax.xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.xaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax.yaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax.zaxis._axinfo["grid"]['color'] = (1,1,1,0)
grad_1_data = gradient_1_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
grad_2_data = gradient_2_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
grad_3_data = gradient_3_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
colors = hcp_myelin_map_left_fsaverage.agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
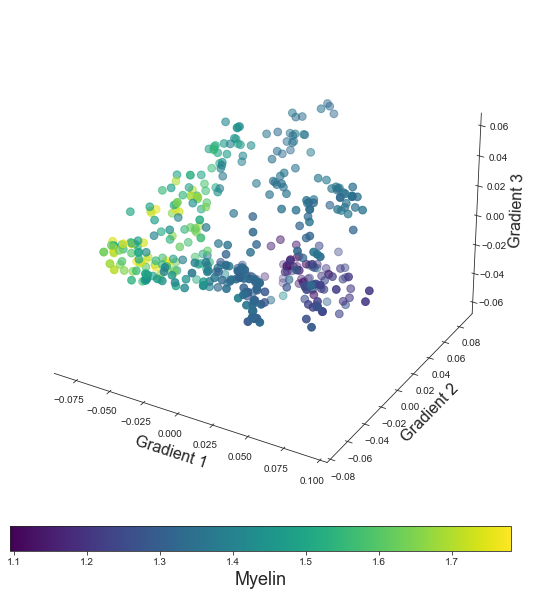
points = ax.scatter3D(grad_1_data, grad_2_data, grad_3_data,
c=colors, cmap='viridis', s=60)
cbar=fig.colorbar(points, orientation='horizontal',
fraction=0.046, pad=0.06, aspect=20)
cbar.set_label('Myelin', fontsize=18)
ax.set_xlabel('Gradient 1', fontsize=16)
ax.set_ylabel('Gradient 2', fontsize=16)
ax.set_zlabel('Gradient 3', fontsize=16)
Text(0.5, 0, 'Gradient 3')
# plot map data (ie feature expression per vertex) in space spanned by the gradients - right hemisphere
sns.set_style('white')
plt.rcParams["font.family"] = "Arial"
fig = plt.figure(figsize=(13,10))
ax = plt.axes(projection="3d")
ax.xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.xaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax.yaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax.zaxis._axinfo["grid"]['color'] = (1,1,1,0)
grad_1_data = gradient_1_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
grad_2_data = gradient_2_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
grad_3_data = gradient_3_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
colors = hcp_myelin_map_right_fsaverage.agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
ax.view_init(27, -120)
ax.invert_xaxis()
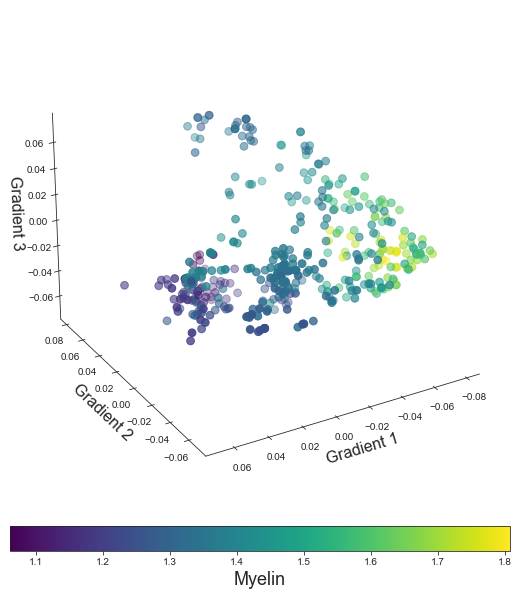
points = ax.scatter3D(grad_1_data, grad_2_data, grad_3_data,
c=colors, cmap='viridis', s=60)
cbar=fig.colorbar(points, orientation='horizontal',
fraction=0.046, pad=0.06, aspect=20)
cbar.set_label('Myelin', fontsize=18)
ax.set_xlabel('Gradient 1', fontsize=16)
ax.set_ylabel('Gradient 2', fontsize=16)
ax.set_zlabel('Gradient 3', fontsize=16)
Text(0.5, 0, 'Gradient 3')
def plot_3d_gradients(feature_left, feature_right, title='title', feature='feature', cmap='viridis'):
# set style & font
sns.set_style('white')
plt.rcParams["font.family"] = "Arial"
# initiate figure
fig = plt.figure(figsize=(17,10))
# initiate first axis, ie left plot
ax1 = fig.add_subplot(121, projection='3d')
# set pane & grid color of first axis
ax1.xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax1.yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax1.zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax1.xaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax1.yaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax1.zaxis._axinfo["grid"]['color'] = (1,1,1,0)
# get data points from three neurosynth ac gradient maps within ac mask - left hemisphere
grad_1_data_left = gradient_1_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
grad_2_data_left = gradient_2_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
grad_3_data_left = gradient_3_surface[0].agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
# get data, ie feature, expression from annotation maps within ac mask - left hemisphere
colors_left = feature_left.agg_data()[np.where(ac_mask_surface[0].agg_data() == 1)]
# create a 3D scatterplot based on neurosynth ac gradient maps data points within ac mask
# and color based on data value in annotation map
points_left = ax1.scatter3D(grad_1_data_left, grad_2_data_left, grad_3_data_left,
c=colors_left, cmap=cmap, s=60)
#cbar=fig.colorbar(points, orientation='horizontal',
# fraction=0.046, pad=0.06, aspect=20)
# set title for first axis, ie left plot
ax1.set_title('left hemisphere', fontsize=18, pad=-40)
# set axis labels
ax1.set_xlabel('Gradient 1', fontsize=16)
ax1.set_ylabel('Gradient 2', fontsize=16)
ax1.set_zlabel('Gradient 3', fontsize=16)
# initiate second axis, ie right plot
ax2 = fig.add_subplot(122, projection='3d')
# set pane & grid color of first axis
ax2.xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax2.yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax2.zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax2.xaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax2.yaxis._axinfo["grid"]['color'] = (1,1,1,0)
ax2.zaxis._axinfo["grid"]['color'] = (1,1,1,0)
# get data points from three neurosynth ac gradient maps within ac mask - right hemisphere
grad_1_data_right = gradient_1_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
grad_2_data_right = gradient_2_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
grad_3_data_right = gradient_3_surface[1].agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
# get data, ie feature, expression from annotation maps within ac mask - right hemisphere
colors_right = feature_right.agg_data()[np.where(ac_mask_surface[1].agg_data() == 1)]
# turn 3D scatterplot along x axis to mirror left
ax2.view_init(27, -120)
ax2.invert_xaxis()
# create a 3D scatterplot based on neurosynth ac gradient maps data points within ac mask
# and color based on data value in annotation map
points_right = ax2.scatter3D(grad_1_data_right, grad_2_data_right, grad_3_data_right,
c=colors_right, cmap=cmap, s=60)
# set title for second axis, ie right plot
ax2.set_title('right hemisphere', fontsize=18, pad=-40)
# set axis labels
ax2.set_xlabel('Gradient 1', fontsize=16)
ax2.set_ylabel('Gradient 2', fontsize=16)
ax2.set_zlabel('Gradient 3', fontsize=16)
# initiate list to store data values of annotation maps in
left_right_list = colors_left.tolist() + colors_right.tolist()
# normalize colors based on annotation map values in left & right hemisphere
norm = mpl.colors.Normalize(vmin=np.min(left_right_list), vmax=np.max(left_right_list))
# create a colorbar based on normalized colors across left & right hemisphere
cbar=fig.colorbar(mpl.cm.ScalarMappable(norm=norm, cmap=cmap),
orientation='horizontal', fraction=0.046, pad=0.02, aspect=12, ax=fig.get_axes(),
)
# set a label for the colorbar
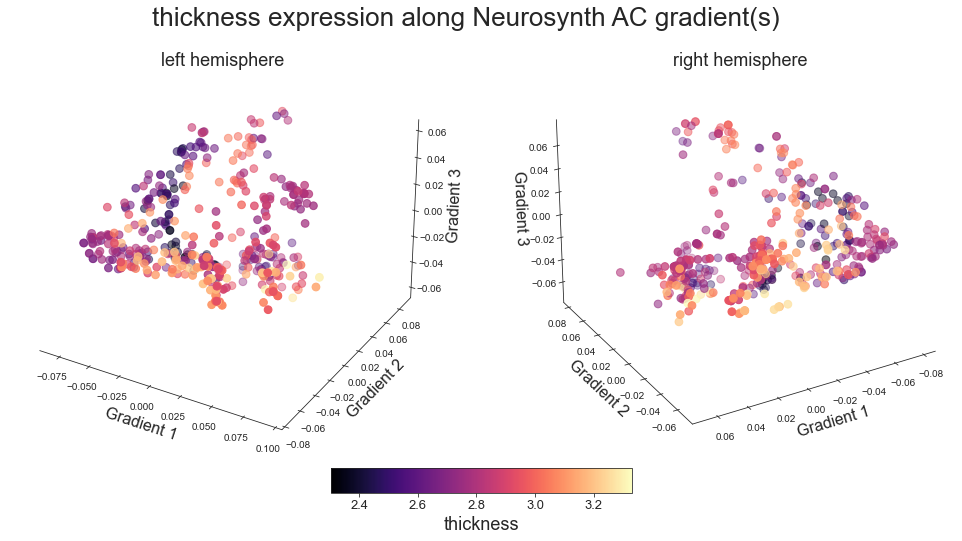
cbar.set_label(feature, fontsize=18)
cbar.ax.tick_params(labelsize=13)
# adapt colorbar: fix missing minimum tick & decrease height
fig.suptitle('%s expression along Neurosynth AC gradient(s)' %title, fontsize=26, y=0.8)
#return left_right_list
plot_3d_gradients(hcp_myelin_map_left_fsaverage, hcp_myelin_map_right_fsaverage, 'myelin', 'myelin (T1w/T2w ratio)', cmap='rocket')
Thickness#
hcp_thickness_maps = datasets.fetch_annotation(source='hcps1200', desc='thickness')
hcp_thickness_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_thickness_maps[0], '10k', hemi='L')[0]
hcp_thickness_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_thickness_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_thickness_map_left_fsaverage, hcp_thickness_map_right_fsaverage), 'fsaverage', '10k', cmap='rocket');
hcp_thickness_map_left_fsaverage_mask_ac, hcp_thickness_map_left_fsaverage_mask_ac_data = mask_surface(hcp_thickness_map_left_fsaverage, ac_mask_surface[0])
hcp_thickness_map_right_fsaverage_mask_ac, hcp_thickness_map_right_fsaverage_mask_ac_data = mask_surface(hcp_thickness_map_right_fsaverage, ac_mask_surface[1])
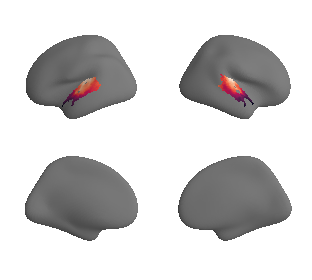
plot_surf_template((hcp_thickness_map_left_fsaverage_mask_ac, hcp_thickness_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='rocket');

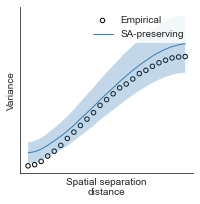
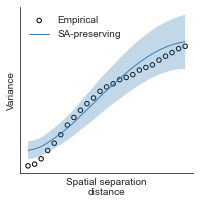
compute_variograms(hcp_thickness_map_left_fsaverage_mask_ac_data, hcp_thickness_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
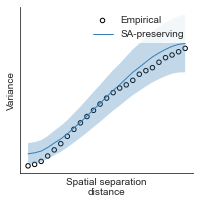
Computing variogram for right hemisphere
hcp_thickness_map_left_fsaverage_mask_ac_surrogates, hcp_thickness_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_thickness_map_left_fsaverage_mask_ac_data,
hcp_thickness_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
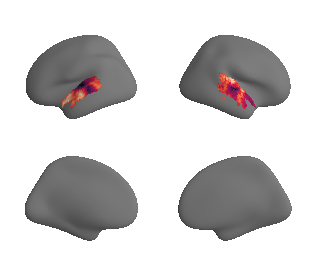
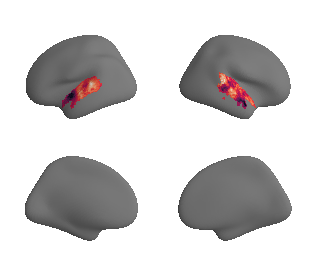
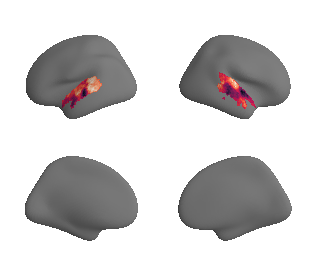
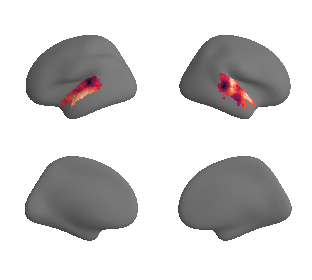
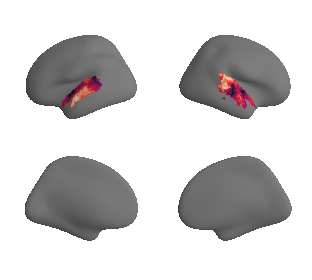
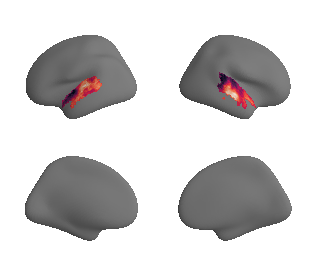
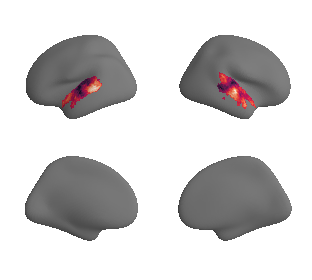
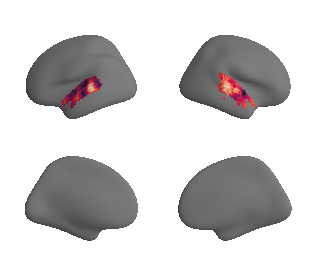
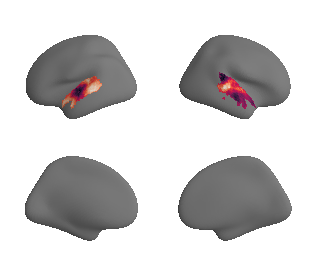
plot_surrogate_maps(hcp_thickness_map_left_fsaverage_mask_ac_surrogates,
hcp_thickness_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='rocket')

grad_hcp_thickness_cors = compare_brain_maps_surrogates(hcp_thickness_map_left_fsaverage_mask_ac_data, hcp_thickness_map_right_fsaverage_mask_ac_data,
hcp_thickness_map_left_fsaverage_mask_ac_surrogates, hcp_thickness_map_right_fsaverage_mask_ac_surrogates,
feature='thickness',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_hcp_thickness_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & thickness are correlated with an r of 0.4470291675571461 at p = 0.171
Working on gradient 2
The LH maps of gradient 2 & thickness are correlated with an r of -0.3748064369066039 at p = 0.237
Working on gradient 3
The LH maps of gradient 3 & thickness are correlated with an r of 0.12424006651557425 at p = 0.751
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & thickness are correlated with an r of 0.3430028992924918 at p = 0.296
Working on gradient 2
The RH maps of gradient 2 & thickness are correlated with an r of -0.41897801251062994 at p = 0.134
Working on gradient 3
The RH maps of gradient 3 & thickness are correlated with an r of 0.28555971648946343 at p = 0.487
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_hcp_thickness_cors.json
plot_3d_gradients(hcp_thickness_map_left_fsaverage, hcp_thickness_map_right_fsaverage, 'thickness', 'thickness', cmap='magma')
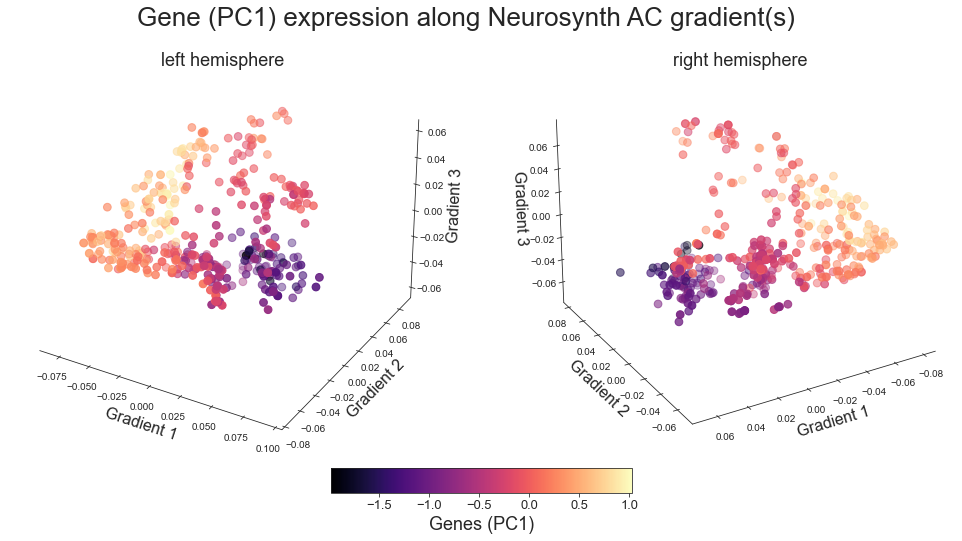
PC 1 genes (based on Allen dataset)#
genepc1_maps = datasets.fetch_annotation(source='abagen', desc='genepc1')
plot_surf_template((genepc1_maps[0], genepc1_maps[1]), 'fsaverage', '10k', cmap='rocket');
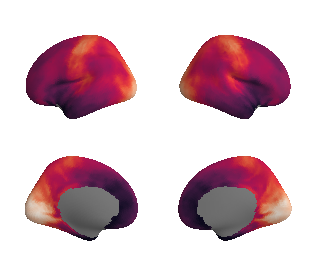
genepc1_map_left_fsaverage_mask_ac, genepc1_map_left_fsaverage_mask_ac_data = mask_surface(genepc1_maps[0], ac_mask_surface[0])
genepc1_map_right_fsaverage_mask_ac, genepc1_map_right_fsaverage_mask_ac_data = mask_surface(genepc1_maps[1], ac_mask_surface[1])
plot_surf_template((genepc1_map_left_fsaverage_mask_ac, genepc1_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='rocket');
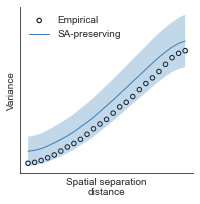
compute_variograms(genepc1_map_left_fsaverage_mask_ac_data, genepc1_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
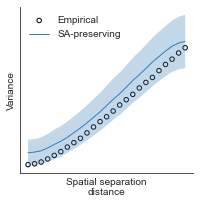
Computing variogram for right hemisphere
genepc1_map_left_fsaverage_mask_ac_surrogates, genepc1_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(genepc1_map_left_fsaverage_mask_ac_data,
genepc1_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
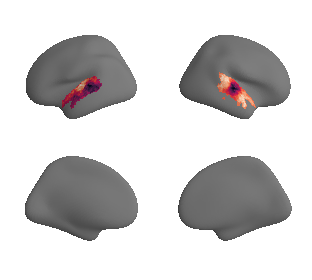
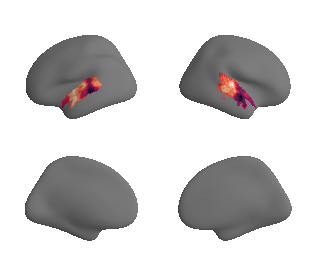
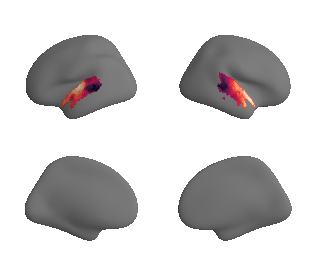
plot_surrogate_maps(genepc1_map_left_fsaverage_mask_ac_surrogates,
genepc1_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='rocket')

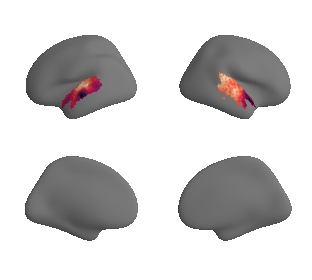
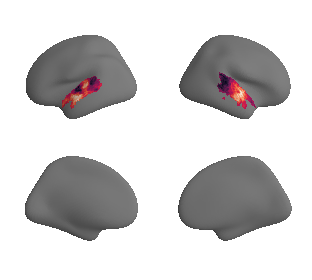
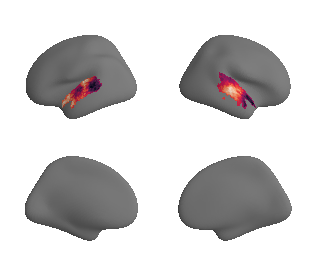
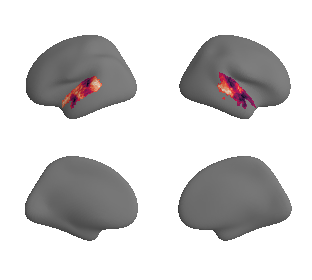
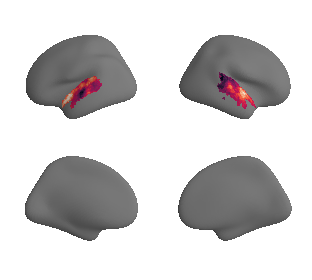
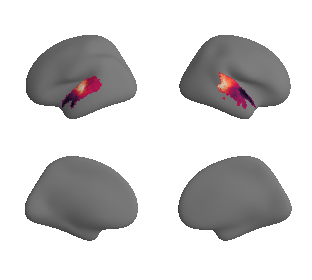
grad_genepc1_cors = compare_brain_maps_surrogates(genepc1_map_left_fsaverage_mask_ac_data, genepc1_map_right_fsaverage_mask_ac_data,
genepc1_map_left_fsaverage_mask_ac_surrogates, genepc1_map_right_fsaverage_mask_ac_surrogates,
feature='thickness',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_genepc1_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & thickness are correlated with an r of -0.6811419581490528 at p = 0.026
Working on gradient 2
The LH maps of gradient 2 & thickness are correlated with an r of -0.2215581658361918 at p = 0.608
Working on gradient 3
The LH maps of gradient 3 & thickness are correlated with an r of 0.5025842813439739 at p = 0.295
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & thickness are correlated with an r of -0.7524630645504091 at p = 0.022
Working on gradient 2
The RH maps of gradient 2 & thickness are correlated with an r of -0.21170769460647496 at p = 0.573
Working on gradient 3
The RH maps of gradient 3 & thickness are correlated with an r of 0.4591025178437719 at p = 0.402
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_genepc1_cors.json
plot_3d_gradients(nb.load(genepc1_maps[0]), nb.load(genepc1_maps[1]), 'Gene (PC1)', 'Genes (PC1)', cmap='magma')
metabolism#
Cerebral blood flow#
raichle_cbf_maps = datasets.fetch_annotation(source='raichle', desc='cbf')
raichle_cbf_map_left_fsaverage = transforms.fslr_to_fsaverage(raichle_cbf_maps[0], '10k', hemi='L')[0]
raichle_cbf_map_right_fsaverage = transforms.fslr_to_fsaverage(raichle_cbf_maps[1], '10k', hemi='R')[0]
plot_surf_template((raichle_cbf_map_left_fsaverage, raichle_cbf_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
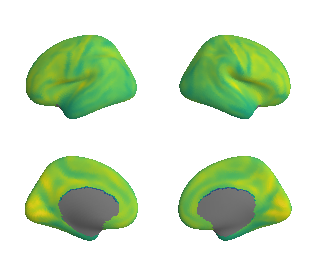
raichle_cbf_map_left_fsaverage_mask_ac, raichle_cbf_map_left_fsaverage_mask_ac_data = mask_surface(raichle_cbf_map_left_fsaverage, ac_mask_surface[0])
raichle_cbf_map_right_fsaverage_mask_ac, raichle_cbf_map_right_fsaverage_mask_ac_data = mask_surface(raichle_cbf_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((raichle_cbf_map_left_fsaverage_mask_ac, raichle_cbf_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
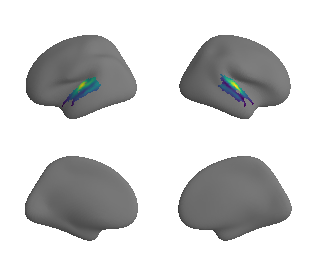
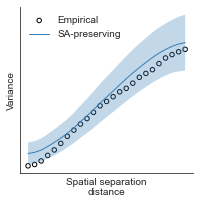
compute_variograms(raichle_cbf_map_left_fsaverage_mask_ac_data, raichle_cbf_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
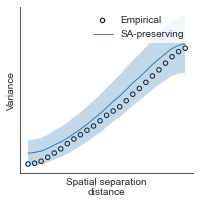
Computing variogram for right hemisphere
raichle_cbf_map_left_fsaverage_mask_ac_surrogates, raichle_cbf_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(raichle_cbf_map_left_fsaverage_mask_ac_data,
raichle_cbf_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
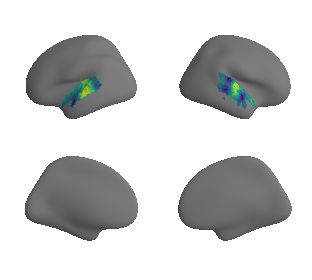
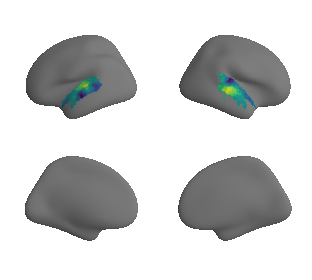
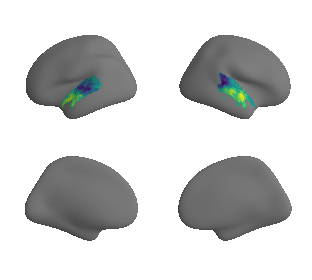
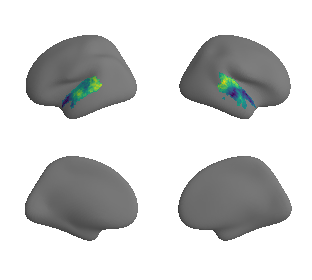
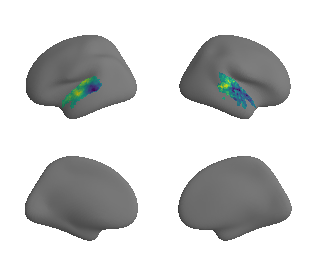
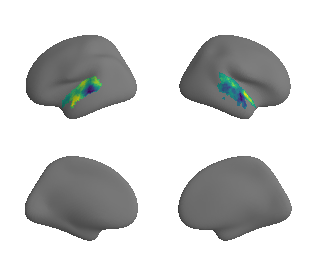
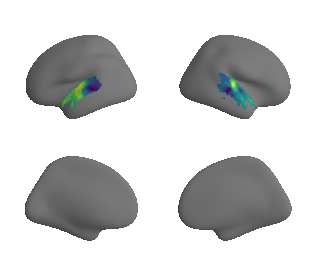
plot_surrogate_maps(raichle_cbf_map_left_fsaverage_mask_ac_surrogates,
raichle_cbf_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='viridis')

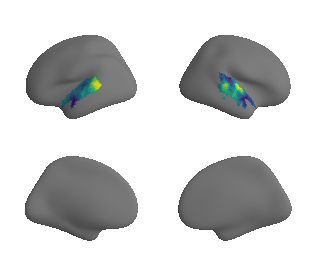

grad_cbf_cors = compare_brain_maps_surrogates(raichle_cbf_map_left_fsaverage_mask_ac_data, raichle_cbf_map_right_fsaverage_mask_ac_data,
raichle_cbf_map_left_fsaverage_mask_ac_surrogates, raichle_cbf_map_right_fsaverage_mask_ac_surrogates,
feature='CBF',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_cbf_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & CBF are correlated with an r of -0.7486988872939675 at p = 0.005
Working on gradient 2
The LH maps of gradient 2 & CBF are correlated with an r of -0.11442495521861 at p = 0.778
Working on gradient 3
The LH maps of gradient 3 & CBF are correlated with an r of 0.23046327083576898 at p = 0.679
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & CBF are correlated with an r of -0.7598222318917669 at p = 0.003
Working on gradient 2
The RH maps of gradient 2 & CBF are correlated with an r of -0.06601923680558318 at p = 0.844
Working on gradient 3
The RH maps of gradient 3 & CBF are correlated with an r of -0.0006462604365873202 at p = 0.999
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_cbf_cors.json
plot_3d_gradients(raichle_cbf_map_left_fsaverage, raichle_cbf_map_right_fsaverage, 'CBF', 'cerebral blood flow', cmap='viridis')
Oxygen#
raichle_oxy_maps = datasets.fetch_annotation(source='raichle', desc='cmr02')
raichle_oxy_map_left_fsaverage = transforms.fslr_to_fsaverage(raichle_oxy_maps[0], '10k', hemi='L')[0]
raichle_oxy_map_right_fsaverage = transforms.fslr_to_fsaverage(raichle_oxy_maps[1], '10k', hemi='R')[0]
plot_surf_template((raichle_oxy_map_left_fsaverage, raichle_oxy_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
raichle_oxy_map_left_fsaverage_mask_ac, raichle_oxy_map_left_fsaverage_mask_ac_data = mask_surface(raichle_oxy_map_left_fsaverage, ac_mask_surface[0])
raichle_oxy_map_right_fsaverage_mask_ac, raichle_oxy_map_right_fsaverage_mask_ac_data = mask_surface(raichle_oxy_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((raichle_oxy_map_left_fsaverage_mask_ac, raichle_oxy_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
compute_variograms(raichle_oxy_map_left_fsaverage_mask_ac_data, raichle_oxy_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
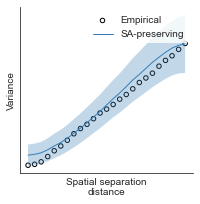
Computing variogram for right hemisphere
raichle_oxy_map_left_fsaverage_mask_ac_surrogates, raichle_oxy_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(raichle_oxy_map_left_fsaverage_mask_ac_data,
raichle_oxy_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
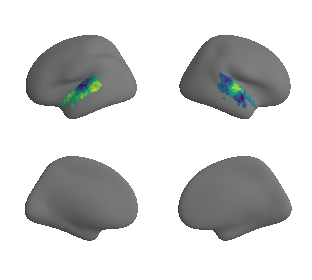
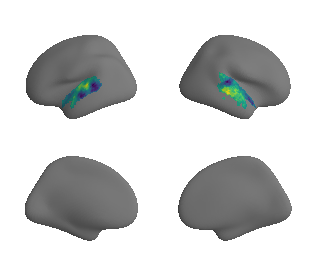
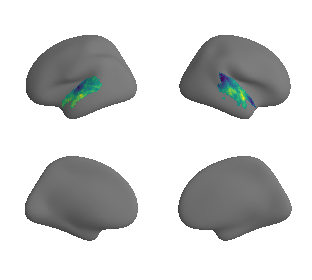
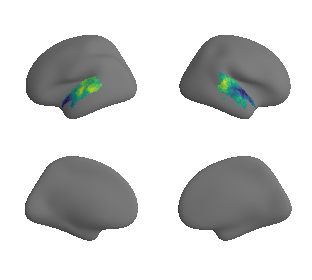
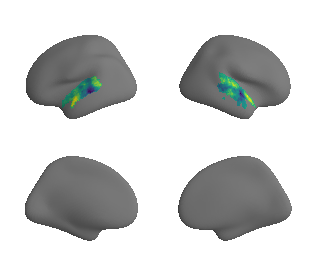
plot_surrogate_maps(raichle_oxy_map_left_fsaverage_mask_ac_surrogates,
raichle_oxy_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='viridis')
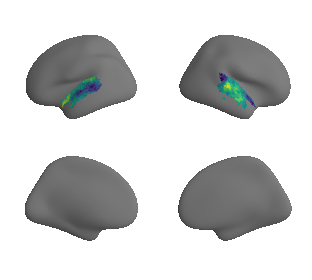
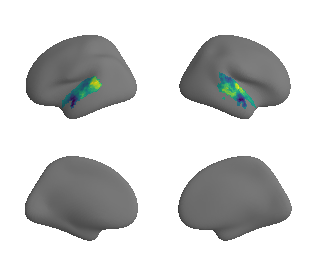


grad_oxy_cors = compare_brain_maps_surrogates(raichle_oxy_map_left_fsaverage_mask_ac_data, raichle_oxy_map_right_fsaverage_mask_ac_data,
raichle_oxy_map_left_fsaverage_mask_ac_surrogates, raichle_oxy_map_right_fsaverage_mask_ac_surrogates,
feature='Oxygen',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_oxy_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Oxygen are correlated with an r of -0.571522938385956 at p = 0.085
Working on gradient 2
The LH maps of gradient 2 & Oxygen are correlated with an r of -0.3444902162944715 at p = 0.355
Working on gradient 3
The LH maps of gradient 3 & Oxygen are correlated with an r of 0.4794579081599584 at p = 0.293
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Oxygen are correlated with an r of -0.6418626491487867 at p = 0.005
Working on gradient 2
The RH maps of gradient 2 & Oxygen are correlated with an r of -0.3269589590743291 at p = 0.258
Working on gradient 3
The RH maps of gradient 3 & Oxygen are correlated with an r of 0.33602519295552075 at p = 0.328
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_oxy_cors.json
plot_3d_gradients(raichle_oxy_map_left_fsaverage, raichle_oxy_map_right_fsaverage, 'Oxygen', 'oxygen', cmap='viridis')
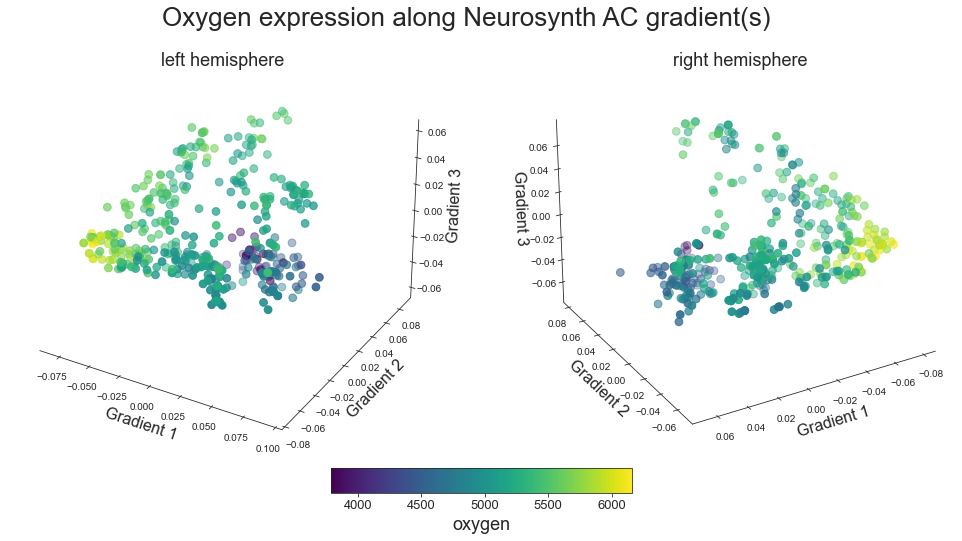
Glucose#
raichle_glu_maps = datasets.fetch_annotation(source='raichle', desc='cmruglu')
raichle_glu_map_left_fsaverage = transforms.fslr_to_fsaverage(raichle_glu_maps[0], '10k', hemi='L')[0]
raichle_glu_map_right_fsaverage = transforms.fslr_to_fsaverage(raichle_glu_maps[1], '10k', hemi='R')[0]
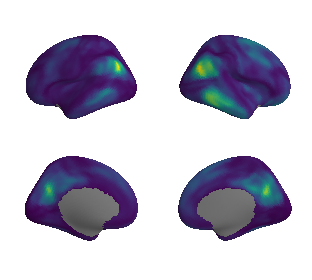
plot_surf_template((raichle_glu_map_left_fsaverage, raichle_glu_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
raichle_glu_map_left_fsaverage_mask_ac, raichle_glu_map_left_fsaverage_mask_ac_data = mask_surface(raichle_glu_map_left_fsaverage, ac_mask_surface[0])
raichle_glu_map_right_fsaverage_mask_ac, raichle_glu_map_right_fsaverage_mask_ac_data = mask_surface(raichle_glu_map_right_fsaverage, ac_mask_surface[1])
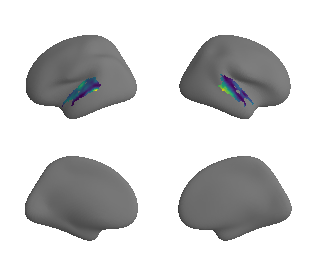
plot_surf_template((raichle_glu_map_left_fsaverage_mask_ac, raichle_glu_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
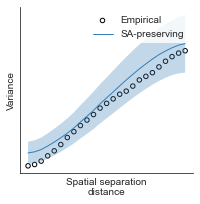
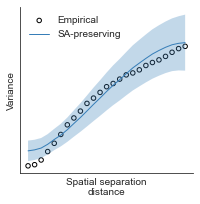
compute_variograms(raichle_glu_map_left_fsaverage_mask_ac_data, raichle_glu_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
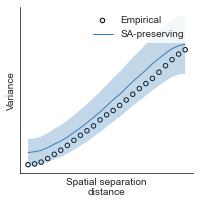
Computing variogram for right hemisphere
raichle_glu_map_left_fsaverage_mask_ac_surrogates, raichle_glu_map_right_fsaverage_mask_ac_data_surrogates = compute_surrogate_maps(raichle_glu_map_left_fsaverage_mask_ac_data,
raichle_glu_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
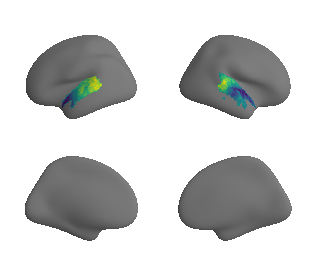
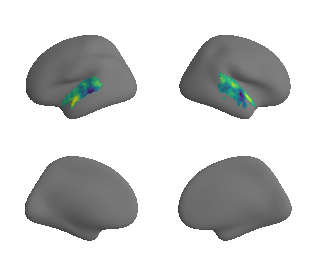
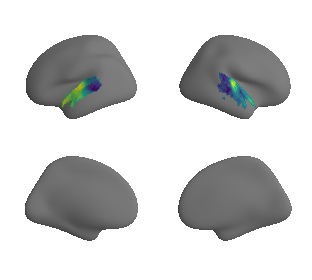
plot_surrogate_maps(raichle_glu_map_left_fsaverage_mask_ac_surrogates,
raichle_glu_map_right_fsaverage_mask_ac_data_surrogates, n_sur=10, cmap='viridis')

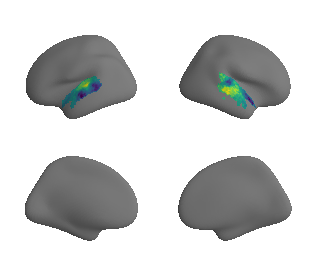
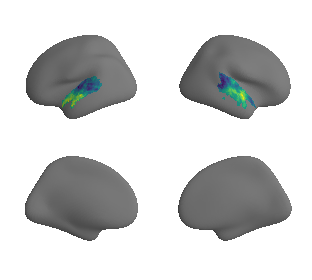

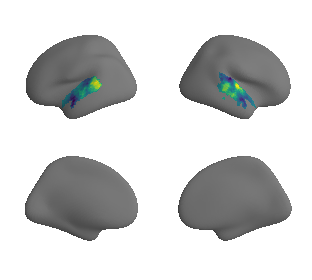


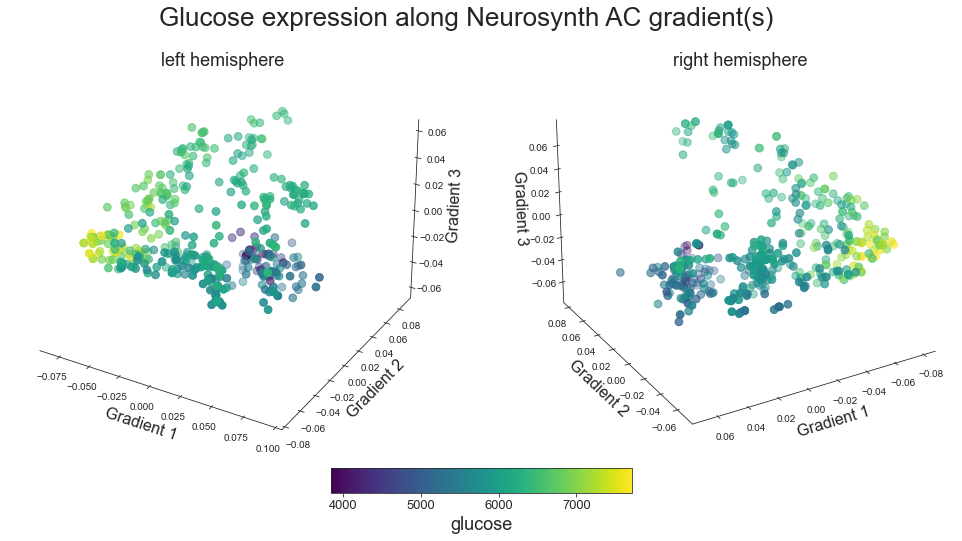
grad_glu_cors = compare_brain_maps_surrogates(raichle_glu_map_left_fsaverage_mask_ac_data, raichle_glu_map_right_fsaverage_mask_ac_data,
raichle_glu_map_left_fsaverage_mask_ac_surrogates, raichle_glu_map_right_fsaverage_mask_ac_data_surrogates,
feature='Glucose',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_glu_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Glucose are correlated with an r of -0.5807375487058407 at p = 0.093
Working on gradient 2
The LH maps of gradient 2 & Glucose are correlated with an r of -0.2948033474057359 at p = 0.457
Working on gradient 3
The LH maps of gradient 3 & Glucose are correlated with an r of 0.45220767170958714 at p = 0.384
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Glucose are correlated with an r of -0.6856478521081981 at p = 0.016
Working on gradient 2
The RH maps of gradient 2 & Glucose are correlated with an r of -0.27689056219940794 at p = 0.382
Working on gradient 3
The RH maps of gradient 3 & Glucose are correlated with an r of 0.32496573507775234 at p = 0.425
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_glu_cors.json
plot_3d_gradients(raichle_glu_map_left_fsaverage, raichle_glu_map_right_fsaverage, 'Glucose', 'glucose', cmap='viridis')
evolution & development#
Evolutionary expansion#
xu_evoexp_maps = datasets.fetch_annotation(source='xu2020', desc='evoexp')
xu_evoexp_map_left_fsaverage = transforms.fslr_to_fsaverage(xu_evoexp_maps[0], '10k', hemi='L')[0]
xu_evoexp_map_right_fsaverage = transforms.fslr_to_fsaverage(xu_evoexp_maps[1], '10k', hemi='R')[0]
plot_surf_template((xu_evoexp_map_left_fsaverage, xu_evoexp_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
xu_evoexp_map_left_fsaverage_mask_ac, xu_evoexp_map_left_fsaverage_mask_ac_data = mask_surface(xu_evoexp_map_left_fsaverage, ac_mask_surface[0])
xu_evoexp_map_right_fsaverage_mask_ac, xu_evoexp_map_right_fsaverage_mask_ac_data = mask_surface(xu_evoexp_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((xu_evoexp_map_left_fsaverage_mask_ac, xu_evoexp_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
compute_variograms(xu_evoexp_map_left_fsaverage_mask_ac_data, xu_evoexp_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
Computing variogram for right hemisphere
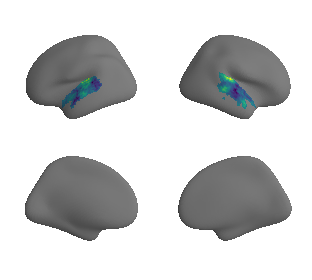
xu_evoexp_map_left_fsaverage_mask_ac_surrogates, xu_evoexp_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(xu_evoexp_map_left_fsaverage_mask_ac_data,
xu_evoexp_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(xu_evoexp_map_left_fsaverage_mask_ac_surrogates,
xu_evoexp_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='viridis')
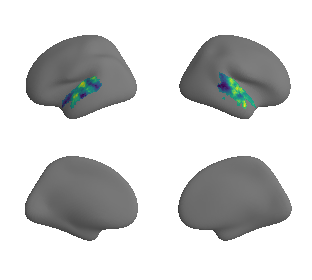

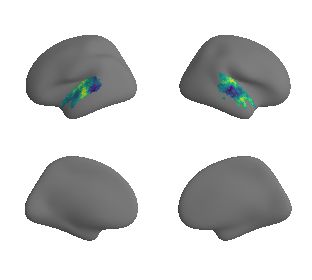

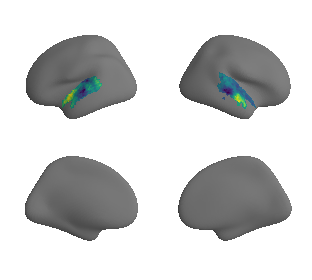
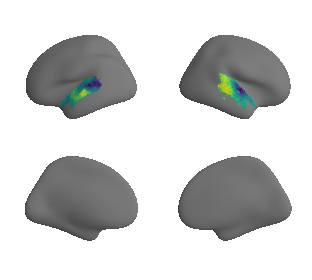

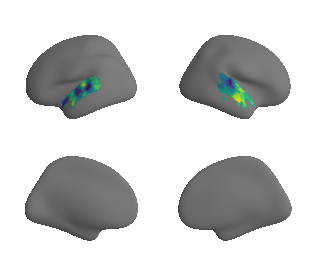
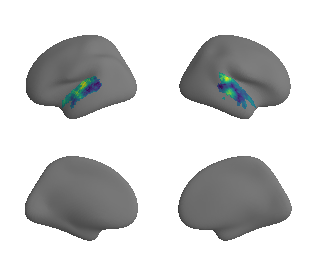
grad_evoexp_cors = compare_brain_maps_surrogates(xu_evoexp_map_left_fsaverage_mask_ac_data, xu_evoexp_map_right_fsaverage_mask_ac_data,
xu_evoexp_map_left_fsaverage_mask_ac_surrogates, xu_evoexp_map_right_fsaverage_mask_ac_surrogates,
feature='Evolutionary expansion',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_evoexp_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Evolutionary expansion are correlated with an r of -0.4191992480272287 at p = 0.14
Working on gradient 2
The LH maps of gradient 2 & Evolutionary expansion are correlated with an r of 0.17483597255835873 at p = 0.581
Working on gradient 3
The LH maps of gradient 3 & Evolutionary expansion are correlated with an r of -0.10142216985911945 at p = 0.773
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Evolutionary expansion are correlated with an r of 0.4310223045748687 at p = 0.25
Working on gradient 2
The RH maps of gradient 2 & Evolutionary expansion are correlated with an r of -0.06140740450543955 at p = 0.859
Working on gradient 3
The RH maps of gradient 3 & Evolutionary expansion are correlated with an r of 0.10830997745429609 at p = 0.834
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_evoexp_cors.json
plot_3d_gradients(xu_evoexp_map_left_fsaverage, xu_evoexp_map_right_fsaverage, 'Evolutionary expansion', 'Evolutionary expansion', cmap='crest_r')
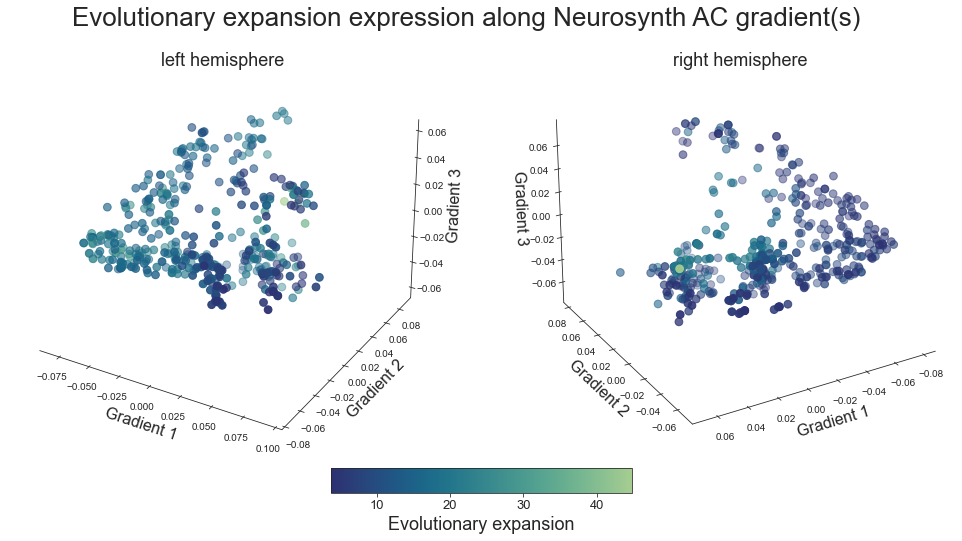
FC homology across species#
xu_fchomology_maps = datasets.fetch_annotation(source='xu2020', desc='FChomology')
xu_fchomology_map_left_fsaverage = transforms.fslr_to_fsaverage(xu_fchomology_maps[0], '10k', hemi='L')[0]
xu_fchomology_map_right_fsaverage = transforms.fslr_to_fsaverage(xu_fchomology_maps[1], '10k', hemi='R')[0]
plot_surf_template((xu_fchomology_map_left_fsaverage, xu_fchomology_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
xu_fchomology_map_left_fsaverage_mask_ac, xu_fchomology_map_left_fsaverage_mask_ac_data = mask_surface(xu_fchomology_map_left_fsaverage, ac_mask_surface[0])
xu_fchomology_map_right_fsaverage_mask_ac, xu_fchomology_map_right_fsaverage_mask_ac_data = mask_surface(xu_fchomology_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((xu_fchomology_map_left_fsaverage_mask_ac, xu_fchomology_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
compute_variograms(xu_fchomology_map_left_fsaverage_mask_ac_data, xu_fchomology_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
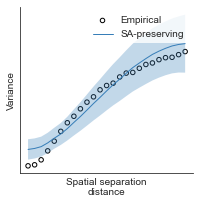
Computing variogram for right hemisphere
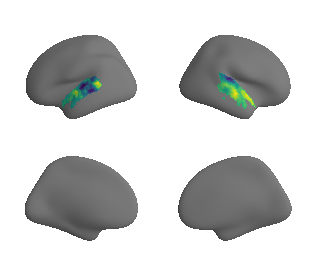
xu_fchomology_map_left_fsaverage_mask_ac_surrogates, xu_fchomology_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(xu_fchomology_map_left_fsaverage_mask_ac_data,
xu_fchomology_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(xu_fchomology_map_left_fsaverage_mask_ac_surrogates,
xu_fchomology_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='viridis')
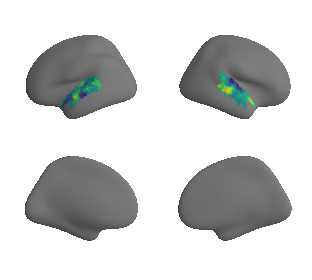

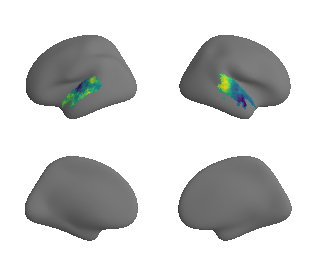

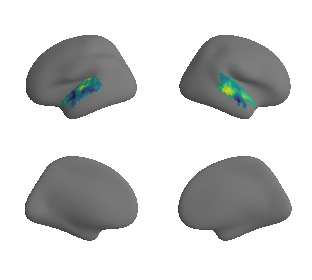

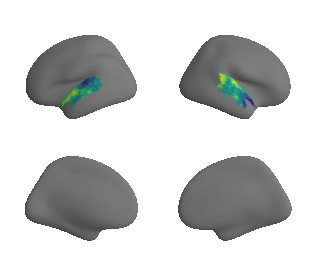
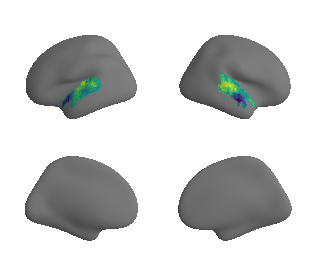
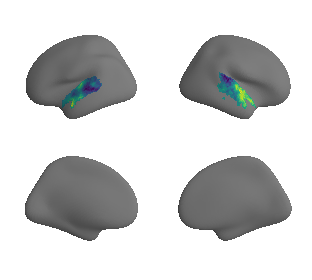
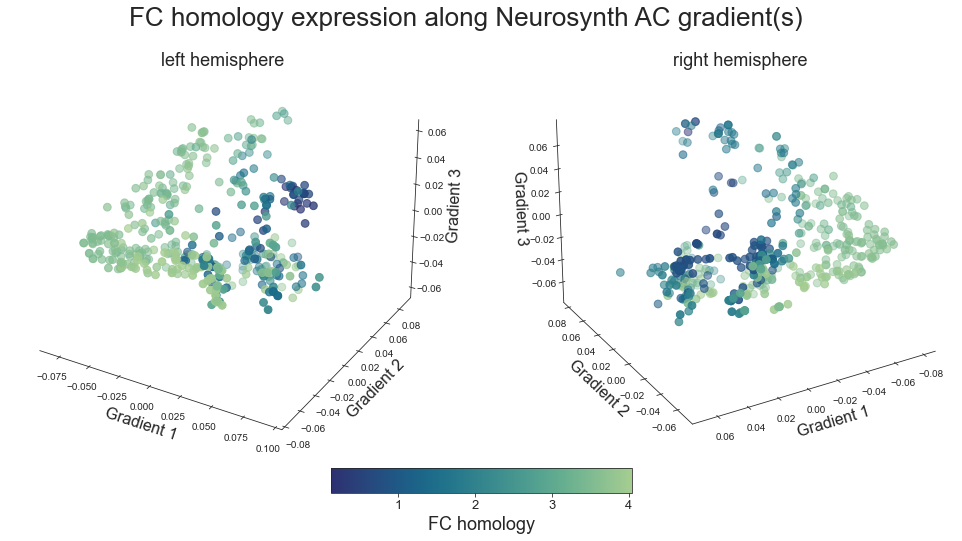
grad_fchomology_cors = compare_brain_maps_surrogates(xu_fchomology_map_left_fsaverage_mask_ac_data, xu_fchomology_map_right_fsaverage_mask_ac_data,
xu_fchomology_map_left_fsaverage_mask_ac_surrogates, xu_fchomology_map_right_fsaverage_mask_ac_surrogates,
feature='FC homology',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_fchomology_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & FC homology are correlated with an r of -0.47810797799604243 at p = 0.068
Working on gradient 2
The LH maps of gradient 2 & FC homology are correlated with an r of -0.369419216817562 at p = 0.218
Working on gradient 3
The LH maps of gradient 3 & FC homology are correlated with an r of -0.012396421377195389 at p = 0.966
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & FC homology are correlated with an r of -0.572902311661132 at p = 0.104
Working on gradient 2
The RH maps of gradient 2 & FC homology are correlated with an r of -0.08402770914836864 at p = 0.785
Working on gradient 3
The RH maps of gradient 3 & FC homology are correlated with an r of -0.41988866379517314 at p = 0.329
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_fchomology_cors.json
plot_3d_gradients(xu_fchomology_map_left_fsaverage, xu_fchomology_map_right_fsaverage, 'FC homology', 'FC homology', cmap='crest_r')
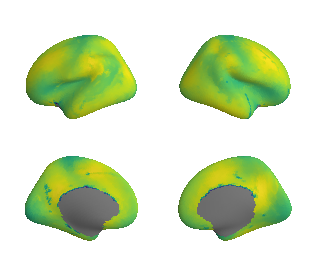
Cortical areal scaling#
reardon_scaling_maps = datasets.fetch_annotation(source='reardon2018', desc='scalinghcp')
reardon_scaling_map_left_fsaverage = transforms.civet_to_fsaverage(reardon_scaling_maps[0], '10k', hemi='L')[0]
reardon_scaling_map_right_fsaverage = transforms.civet_to_fsaverage(reardon_scaling_maps[1], '10k', hemi='R')[0]
plot_surf_template((reardon_scaling_map_left_fsaverage, reardon_scaling_map_right_fsaverage), 'fsaverage', '10k', cmap='viridis');
reardon_scaling_map_left_fsaverage_mask_ac, reardon_scaling_map_left_fsaverage_mask_ac_data = mask_surface(reardon_scaling_map_left_fsaverage, ac_mask_surface[0])
reardon_scaling_map_right_fsaverage_mask_ac, reardon_scaling_map_right_fsaverage_mask_ac_data = mask_surface(reardon_scaling_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((reardon_scaling_map_left_fsaverage_mask_ac, reardon_scaling_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap='viridis');
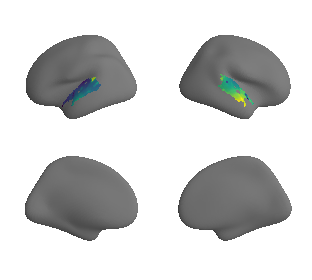
compute_variograms(reardon_scaling_map_left_fsaverage_mask_ac_data, reardon_scaling_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
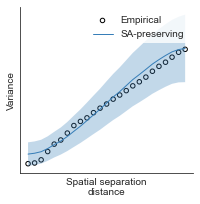
Computing variogram for right hemisphere
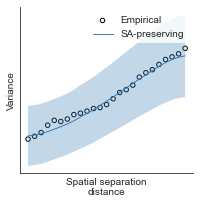
reardon_scaling_map_left_fsaverage_mask_ac_surrogates, reardon_scaling_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(reardon_scaling_map_left_fsaverage_mask_ac_data,
reardon_scaling_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(reardon_scaling_map_left_fsaverage_mask_ac_surrogates,
reardon_scaling_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap='viridis')
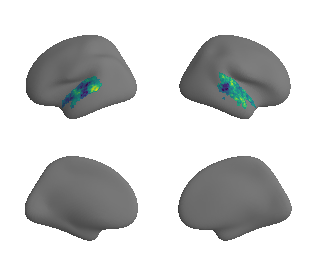
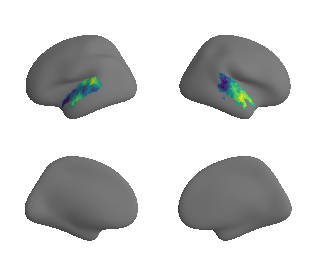
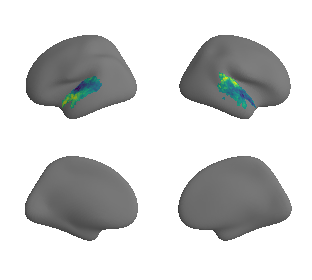
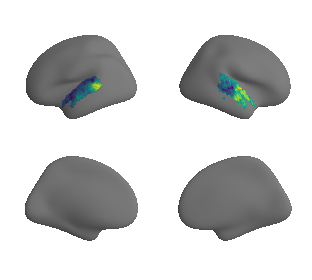
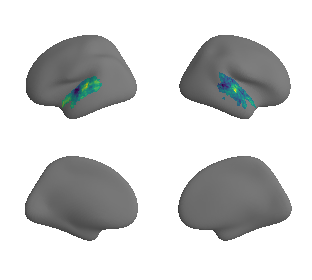
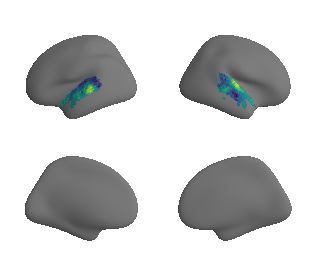
grad_scaling_cors = compare_brain_maps_surrogates(reardon_scaling_map_left_fsaverage_mask_ac_data, reardon_scaling_map_right_fsaverage_mask_ac_data,
reardon_scaling_map_left_fsaverage_mask_ac_surrogates, reardon_scaling_map_right_fsaverage_mask_ac_surrogates,
feature='Cortical scaling',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_corticalscaling_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Cortical scaling are correlated with an r of 0.38589847439113006 at p = 0.233
Working on gradient 2
The LH maps of gradient 2 & Cortical scaling are correlated with an r of 0.021526789277321236 at p = 0.943
Working on gradient 3
The LH maps of gradient 3 & Cortical scaling are correlated with an r of 0.42710008849272985 at p = 0.277
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Cortical scaling are correlated with an r of 0.5985472862068709 at p = 0.055
Working on gradient 2
The RH maps of gradient 2 & Cortical scaling are correlated with an r of -0.0943922789255639 at p = 0.778
Working on gradient 3
The RH maps of gradient 3 & Cortical scaling are correlated with an r of -0.13787432108862127 at p = 0.784
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_corticalscaling_cors.json
plot_3d_gradients(reardon_scaling_map_left_fsaverage, reardon_scaling_map_right_fsaverage, 'Cortical area scaling', 'Cortical area scaling', cmap='crest_r')
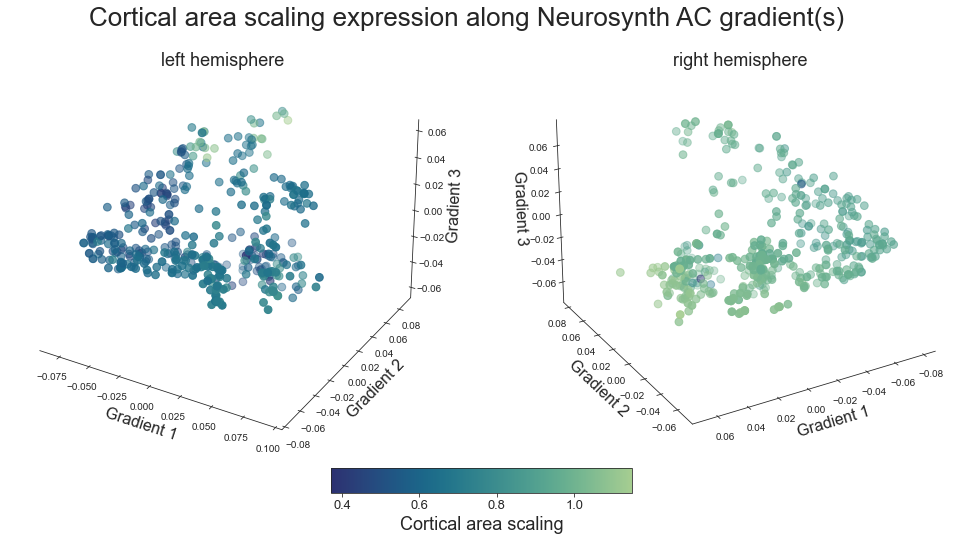
electrophysiology#
MEG - Alpha#
cmap_ephys = sns.cubehelix_palette(as_cmap=True, reverse=True)
hcp_megalpha_maps = datasets.fetch_annotation(source='hcps1200', desc='megalpha')
hcp_megalpha_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_megalpha_maps[0], '10k', hemi='L')[0]
hcp_megalpha_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_megalpha_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_megalpha_map_left_fsaverage, hcp_megalpha_map_right_fsaverage), 'fsaverage', '10k', cmap=cmap_ephys);
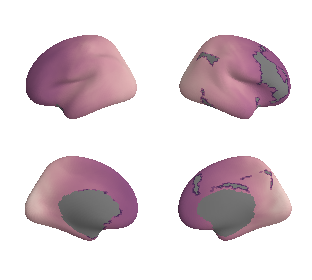
hcp_megalpha_map_left_fsaverage_mask_ac, hcp_megalpha_map_left_fsaverage_mask_ac_data = mask_surface(hcp_megalpha_map_left_fsaverage, ac_mask_surface[0])
hcp_megalpha_map_right_fsaverage_mask_ac, hcp_megalpha_map_right_fsaverage_mask_ac_data = mask_surface(hcp_megalpha_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((hcp_megalpha_map_left_fsaverage_mask_ac, hcp_megalpha_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmap_ephys);
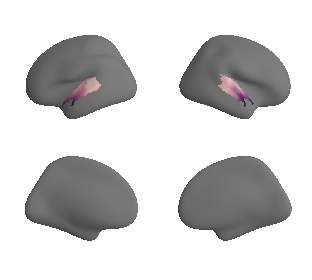
compute_variograms(hcp_megalpha_map_left_fsaverage_mask_ac_data, hcp_megalpha_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
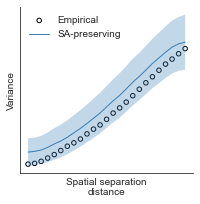
Computing variogram for right hemisphere
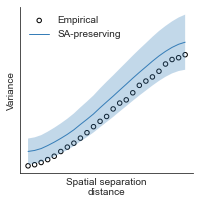
hcp_megalpha_map_left_fsaverage_mask_ac_surrogates, hcp_megalpha_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_megalpha_map_left_fsaverage_mask_ac_data,
hcp_megalpha_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(hcp_megalpha_map_left_fsaverage_mask_ac_surrogates,
hcp_megalpha_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmap_ephys)

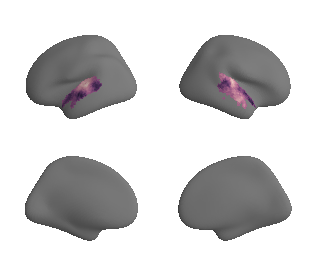
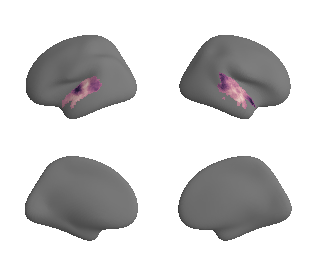
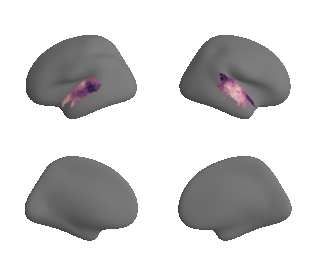
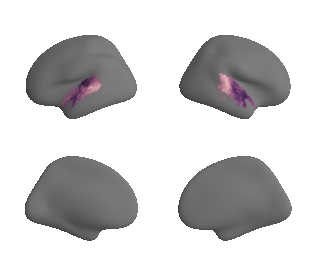

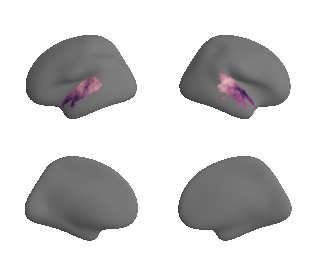
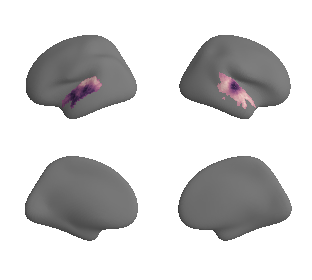
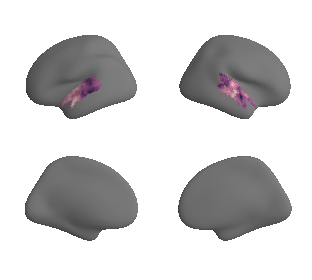
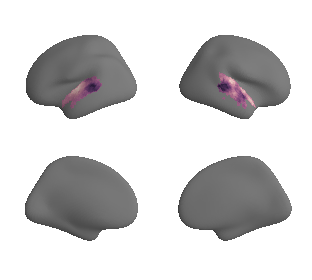
grad_meg_alpha_cors = compare_brain_maps_surrogates(hcp_megalpha_map_left_fsaverage_mask_ac_data, hcp_megalpha_map_right_fsaverage_mask_ac_data,
hcp_megalpha_map_left_fsaverage_mask_ac_surrogates, hcp_megalpha_map_right_fsaverage_mask_ac_surrogates,
feature='MEG-Alpha',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_alpha_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & MEG-Alpha are correlated with an r of -0.3988069984831253 at p = 0.303
Working on gradient 2
The LH maps of gradient 2 & MEG-Alpha are correlated with an r of 0.004798642232330922 at p = 0.993
Working on gradient 3
The LH maps of gradient 3 & MEG-Alpha are correlated with an r of 0.6833848522958194 at p = 0.143
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & MEG-Alpha are correlated with an r of -0.44307446050358584 at p = 0.277
Working on gradient 2
The RH maps of gradient 2 & MEG-Alpha are correlated with an r of -0.10722970486970439 at p = 0.77
Working on gradient 3
The RH maps of gradient 3 & MEG-Alpha are correlated with an r of 0.5712577734124289 at p = 0.197
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_alpha_cors.json
plot_3d_gradients(hcp_megalpha_map_left_fsaverage, hcp_megalpha_map_right_fsaverage, 'MEG - Alpha', 'MEG - Alpha', cmap=sns.cubehelix_palette(as_cmap=True, reverse=True))
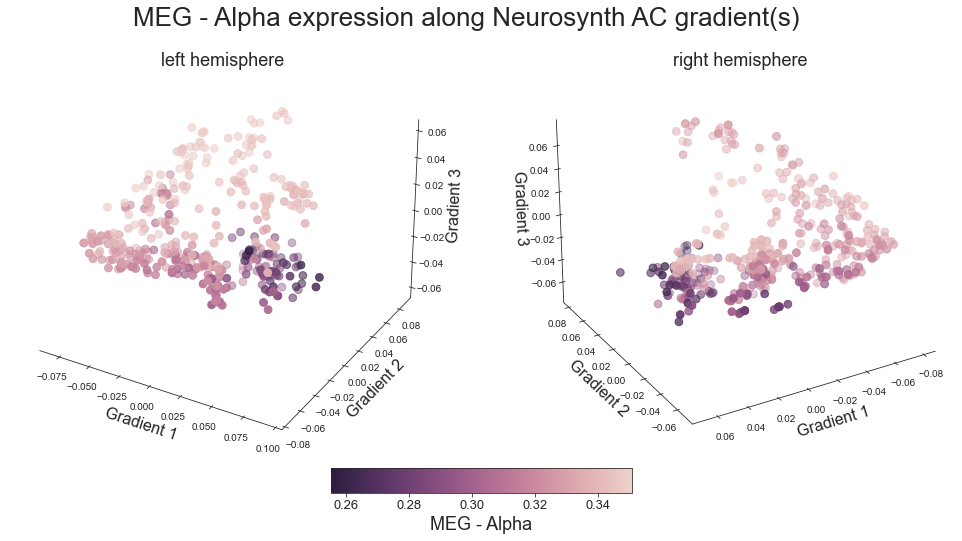
MEG - Beta#
hcp_megbeta_maps = datasets.fetch_annotation(source='hcps1200', desc='megbeta')
hcp_megbeta_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_megbeta_maps[0], '10k', hemi='L')[0]
hcp_megbeta_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_megbeta_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_megbeta_map_left_fsaverage, hcp_megbeta_map_right_fsaverage), 'fsaverage', '10k', cmap=cmap_ephys);
hcp_megbeta_map_left_fsaverage_mask_ac, hcp_megbeta_map_left_fsaverage_mask_ac_data = mask_surface(hcp_megbeta_map_left_fsaverage, ac_mask_surface[0])
hcp_megbeta_map_right_fsaverage_mask_ac, hcp_megbeta_map_right_fsaverage_mask_ac_data = mask_surface(hcp_megbeta_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((hcp_megbeta_map_left_fsaverage_mask_ac, hcp_megbeta_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmap_ephys);
compute_variograms(hcp_megbeta_map_left_fsaverage_mask_ac_data, hcp_megbeta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
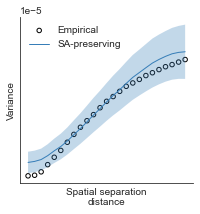
Computing variogram for right hemisphere
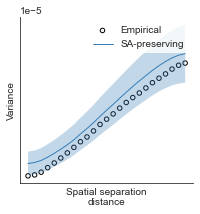
hcp_megbeta_map_left_fsaverage_mask_ac_surrogates, hcp_megbeta_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_megbeta_map_left_fsaverage_mask_ac_data,
hcp_megbeta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(hcp_megbeta_map_left_fsaverage_mask_ac_surrogates,
hcp_megbeta_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmap_ephys)
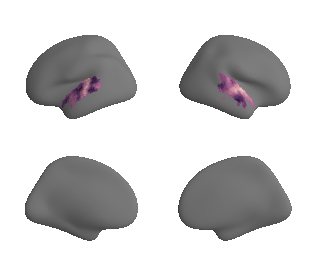
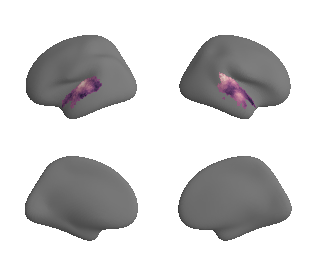
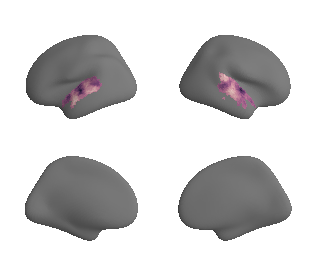
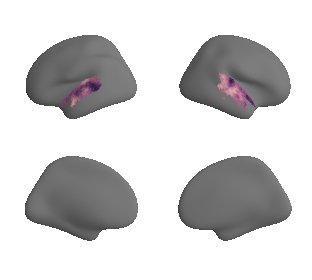
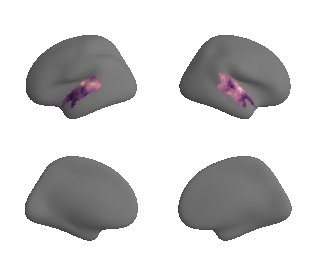
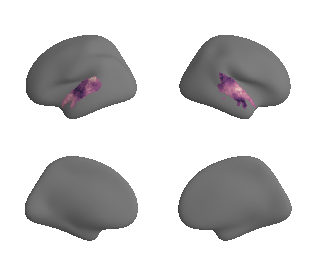
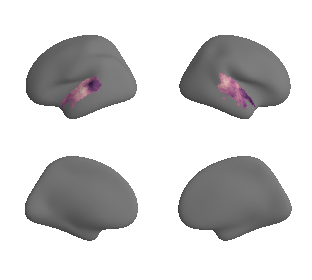
grad_meg_beta_cors = compare_brain_maps_surrogates(hcp_megbeta_map_left_fsaverage_mask_ac_data, hcp_megbeta_map_right_fsaverage_mask_ac_data,
hcp_megbeta_map_left_fsaverage_mask_ac_surrogates, hcp_megbeta_map_right_fsaverage_mask_ac_surrogates,
feature='MEG-Beta',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_beta_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & MEG-Beta are correlated with an r of -0.4123555488988108 at p = 0.157
Working on gradient 2
The LH maps of gradient 2 & MEG-Beta are correlated with an r of 0.27966249666824083 at p = 0.389
Working on gradient 3
The LH maps of gradient 3 & MEG-Beta are correlated with an r of 0.3879766971225199 at p = 0.269
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & MEG-Beta are correlated with an r of -0.4176256193401211 at p = 0.261
Working on gradient 2
The RH maps of gradient 2 & MEG-Beta are correlated with an r of 0.3152128279068916 at p = 0.327
Working on gradient 3
The RH maps of gradient 3 & MEG-Beta are correlated with an r of 0.537051178157867 at p = 0.211
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_beta_cors.json
plot_3d_gradients(hcp_megbeta_map_left_fsaverage, hcp_megbeta_map_right_fsaverage, 'MEG - Beta', 'MEG - Beta', cmap=sns.cubehelix_palette(as_cmap=True, reverse=True))
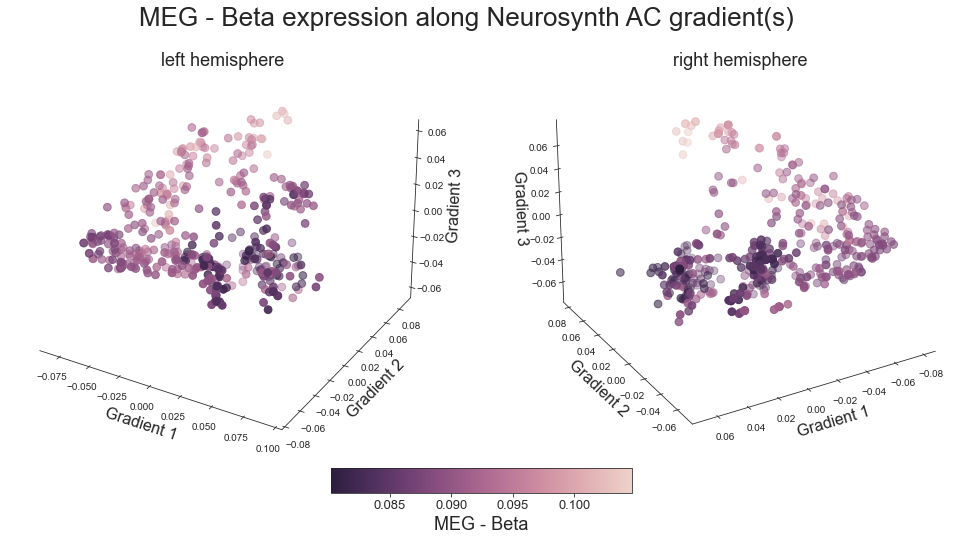
MEG - Delta#
hcp_megdelta_maps = datasets.fetch_annotation(source='hcps1200', desc='megdelta')
hcp_megdelta_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_megdelta_maps[0], '10k', hemi='L')[0]
hcp_megdelta_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_megdelta_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_megdelta_map_left_fsaverage, hcp_megdelta_map_right_fsaverage), 'fsaverage', '10k', cmap=cmap_ephys);

hcp_megdelta_map_left_fsaverage_mask_ac, hcp_megdelta_map_left_fsaverage_mask_ac_data = mask_surface(hcp_megdelta_map_left_fsaverage, ac_mask_surface[0])
hcp_megdelta_map_right_fsaverage_mask_ac, hcp_megdelta_map_right_fsaverage_mask_ac_data = mask_surface(hcp_megdelta_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((hcp_megdelta_map_left_fsaverage_mask_ac, hcp_megdelta_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmap_ephys);
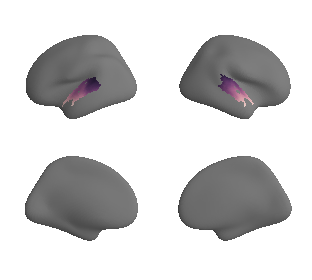
compute_variograms(hcp_megdelta_map_left_fsaverage_mask_ac_data, hcp_megdelta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
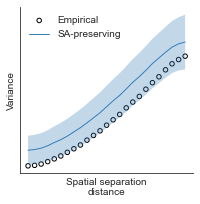
Computing variogram for right hemisphere
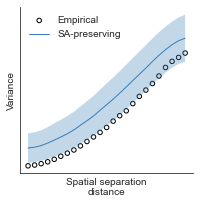
hcp_megdelta_map_left_fsaverage_mask_ac_surrogates, hcp_megdelta_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_megdelta_map_left_fsaverage_mask_ac_data,
hcp_megdelta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(hcp_megdelta_map_left_fsaverage_mask_ac_surrogates,
hcp_megdelta_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmap_ephys)
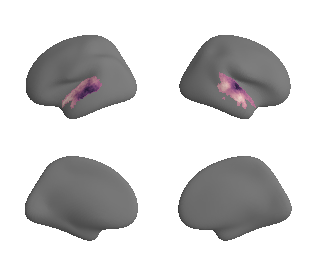
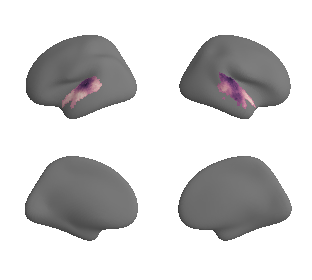
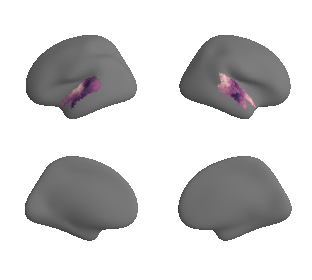
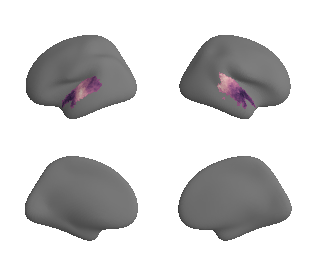
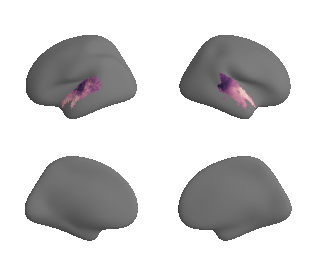
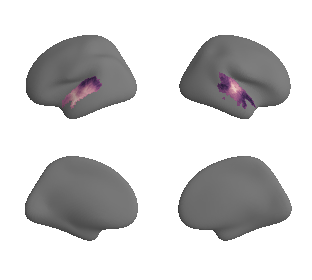
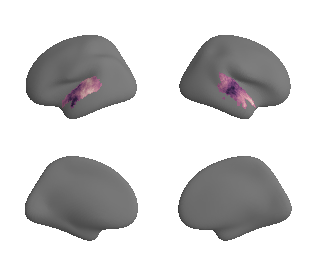
grad_meg_delta_cors = compare_brain_maps_surrogates(hcp_megdelta_map_left_fsaverage_mask_ac_data, hcp_megdelta_map_right_fsaverage_mask_ac_data,
hcp_megdelta_map_left_fsaverage_mask_ac_surrogates, hcp_megdelta_map_right_fsaverage_mask_ac_surrogates,
feature='MEG-Delta',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_delta_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & MEG-Delta are correlated with an r of 0.38472630707022787 at p = 0.33
Working on gradient 2
The LH maps of gradient 2 & MEG-Delta are correlated with an r of -0.08028735020913765 at p = 0.868
Working on gradient 3
The LH maps of gradient 3 & MEG-Delta are correlated with an r of -0.7605364781146876 at p = 0.067
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & MEG-Delta are correlated with an r of 0.5070833831283196 at p = 0.237
Working on gradient 2
The RH maps of gradient 2 & MEG-Delta are correlated with an r of -0.04615872469818411 at p = 0.901
Working on gradient 3
The RH maps of gradient 3 & MEG-Delta are correlated with an r of -0.6992796111693914 at p = 0.146
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_delta_cors.json
plot_3d_gradients(hcp_megdelta_map_left_fsaverage, hcp_megdelta_map_right_fsaverage, 'MEG - Delta', 'MEG - Delta', cmap=sns.cubehelix_palette(as_cmap=True, reverse=True))
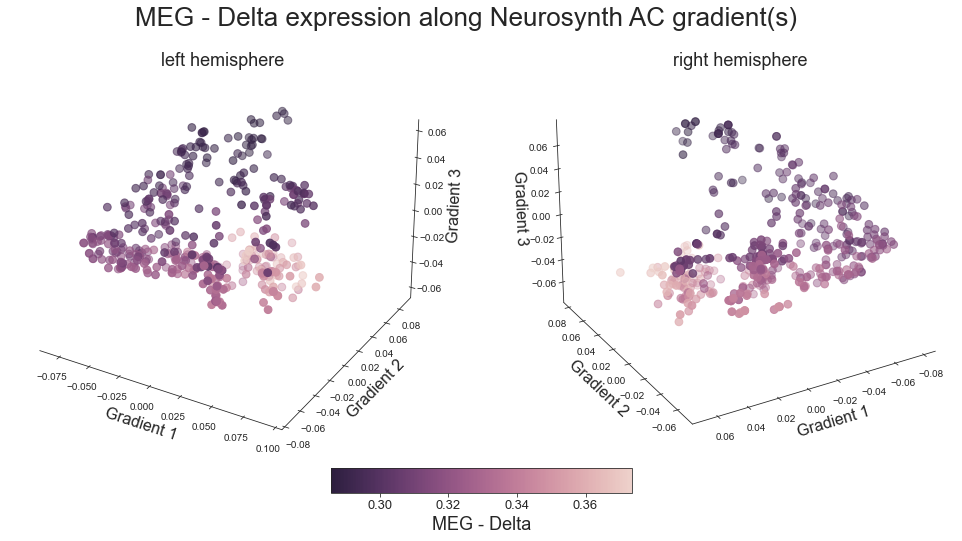
MEG - Theta#
hcp_megtheta_maps = datasets.fetch_annotation(source='hcps1200', desc='megtheta')
hcp_megtheta_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_megtheta_maps[0], '10k', hemi='L')[0]
hcp_megtheta_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_megtheta_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_megtheta_map_left_fsaverage, hcp_megtheta_map_right_fsaverage), 'fsaverage', '10k', cmap=cmap_ephys);

hcp_megtheta_map_left_fsaverage_mask_ac, hcp_megtheta_map_left_fsaverage_mask_ac_data = mask_surface(hcp_megtheta_map_left_fsaverage, ac_mask_surface[0])
hcp_megtheta_map_right_fsaverage_mask_ac, hcp_megtheta_map_right_fsaverage_mask_ac_data = mask_surface(hcp_megtheta_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((hcp_megtheta_map_left_fsaverage_mask_ac, hcp_megtheta_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmap_ephys);
compute_variograms(hcp_megtheta_map_left_fsaverage_mask_ac_data, hcp_megtheta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
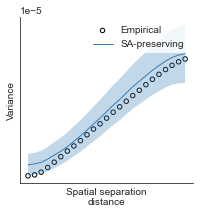
Computing variogram for right hemisphere
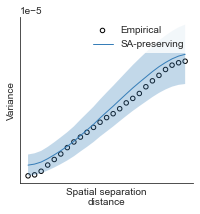
hcp_megtheta_map_left_fsaverage_mask_ac_surrogates, hcp_megtheta_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_megtheta_map_left_fsaverage_mask_ac_data,
hcp_megtheta_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(hcp_megtheta_map_left_fsaverage_mask_ac_surrogates,
hcp_megtheta_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmap_ephys)
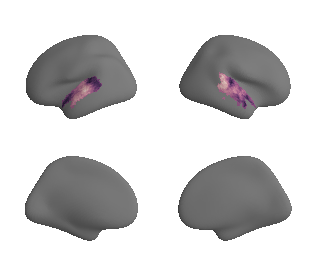
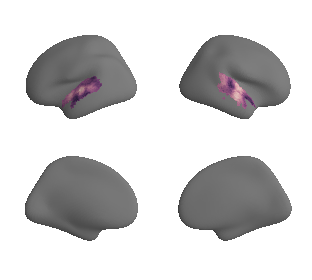
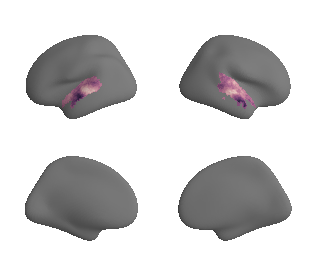
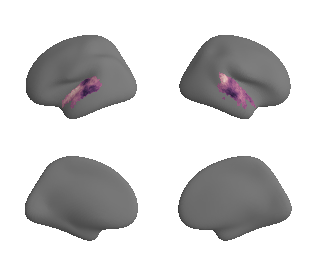
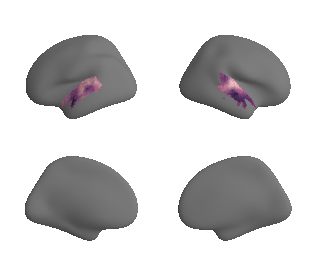
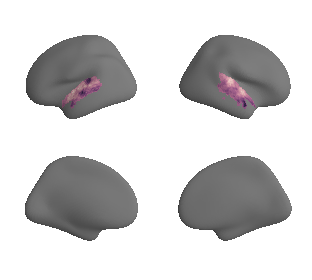
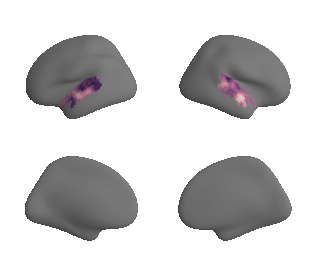
grad_meg_theta_cors = compare_brain_maps_surrogates(hcp_megtheta_map_left_fsaverage_mask_ac_data, hcp_megtheta_map_right_fsaverage_mask_ac_data,
hcp_megtheta_map_left_fsaverage_mask_ac_surrogates, hcp_megtheta_map_right_fsaverage_mask_ac_surrogates,
feature='MEG-Theta',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_theta_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & MEG-Theta are correlated with an r of -0.05993636580464229 at p = 0.876
Working on gradient 2
The LH maps of gradient 2 & MEG-Theta are correlated with an r of 0.31467466677919903 at p = 0.374
Working on gradient 3
The LH maps of gradient 3 & MEG-Theta are correlated with an r of 0.3133905730916277 at p = 0.478
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & MEG-Theta are correlated with an r of -0.33514758353924556 at p = 0.368
Working on gradient 2
The RH maps of gradient 2 & MEG-Theta are correlated with an r of 0.5014872973932596 at p = 0.095
Working on gradient 3
The RH maps of gradient 3 & MEG-Theta are correlated with an r of 0.06928691918939793 at p = 0.87
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_theta_cors.json
plot_3d_gradients(hcp_megtheta_map_left_fsaverage, hcp_megtheta_map_right_fsaverage, 'MEG - Theta', 'MEG - Theta', cmap=sns.cubehelix_palette(as_cmap=True, reverse=True))
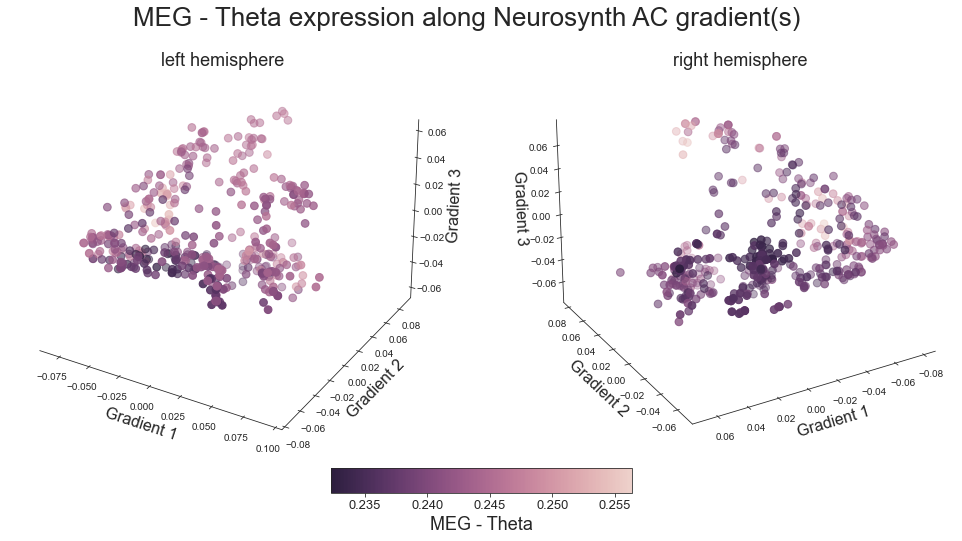
MEG - Intrinsic timescale#
hcp_timescale_maps = datasets.fetch_annotation(source='hcps1200', desc='megtimescale')
hcp_timescale_map_left_fsaverage = transforms.fslr_to_fsaverage(hcp_timescale_maps[0], '10k', hemi='L')[0]
hcp_timescale_map_right_fsaverage = transforms.fslr_to_fsaverage(hcp_timescale_maps[1], '10k', hemi='R')[0]
plot_surf_template((hcp_timescale_map_left_fsaverage, hcp_timescale_map_right_fsaverage), 'fsaverage', '10k', cmap=cmap_ephys);
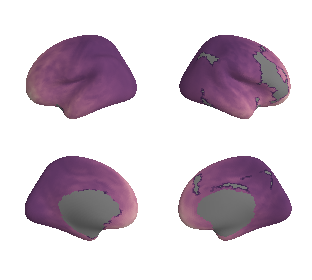
hcp_timescale_map_left_fsaverage_mask_ac, hcp_timescale_map_left_fsaverage_mask_ac_data = mask_surface(hcp_timescale_map_left_fsaverage, ac_mask_surface[0])
hcp_timescale_map_right_fsaverage_mask_ac, hcp_timescale_map_right_fsaverage_mask_ac_data = mask_surface(hcp_timescale_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((hcp_timescale_map_left_fsaverage_mask_ac, hcp_timescale_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmap_ephys);
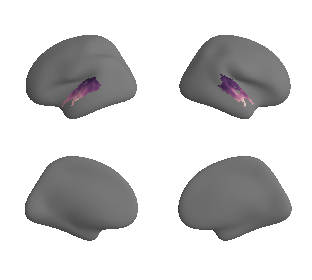
compute_variograms(hcp_timescale_map_left_fsaverage_mask_ac_data, hcp_timescale_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
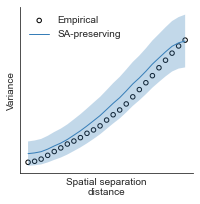
Computing variogram for right hemisphere
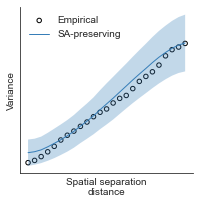
hcp_timescale_map_left_fsaverage_mask_ac_surrogates, hcp_timescale_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(hcp_timescale_map_left_fsaverage_mask_ac_data,
hcp_timescale_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(hcp_timescale_map_left_fsaverage_mask_ac_surrogates,
hcp_timescale_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmap_ephys)
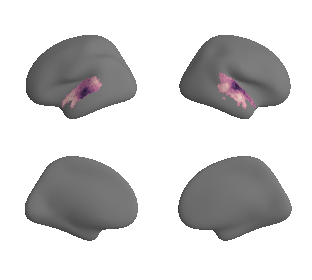
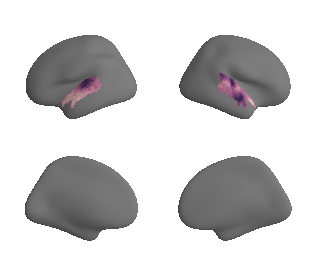
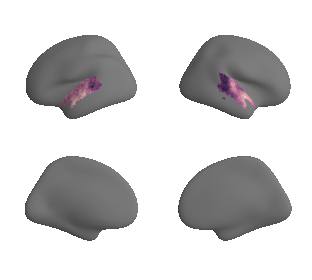
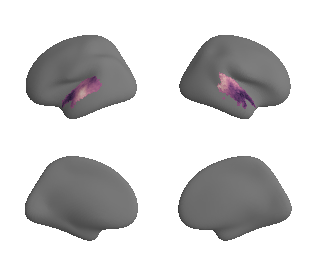
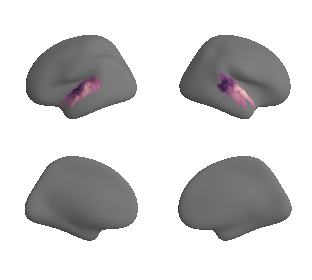
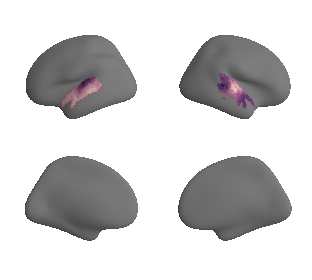
grad_meg_timescale_cors = compare_brain_maps_surrogates(hcp_timescale_map_left_fsaverage_mask_ac_data, hcp_timescale_map_right_fsaverage_mask_ac_data,
hcp_timescale_map_left_fsaverage_mask_ac_surrogates, hcp_timescale_map_right_fsaverage_mask_ac_surrogates,
feature='MEG-Intrinsic Timescale',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_timescale_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & MEG-Intrinsic Timescale are correlated with an r of 0.5995167264884204 at p = 0.102
Working on gradient 2
The LH maps of gradient 2 & MEG-Intrinsic Timescale are correlated with an r of -0.15830945857225875 at p = 0.727
Working on gradient 3
The LH maps of gradient 3 & MEG-Intrinsic Timescale are correlated with an r of -0.5483853242797996 at p = 0.315
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & MEG-Intrinsic Timescale are correlated with an r of 0.6031668431713991 at p = 0.087
Working on gradient 2
The RH maps of gradient 2 & MEG-Intrinsic Timescale are correlated with an r of -0.026940688553277345 at p = 0.924
Working on gradient 3
The RH maps of gradient 3 & MEG-Intrinsic Timescale are correlated with an r of -0.5485926442736488 at p = 0.243
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_meg_timescale_cors.json
plot_3d_gradients(hcp_timescale_map_left_fsaverage, hcp_timescale_map_right_fsaverage, 'MEG - intrinsic timescale', 'MEG - intrinsic timescale', cmap=sns.cubehelix_palette(as_cmap=True, reverse=True))
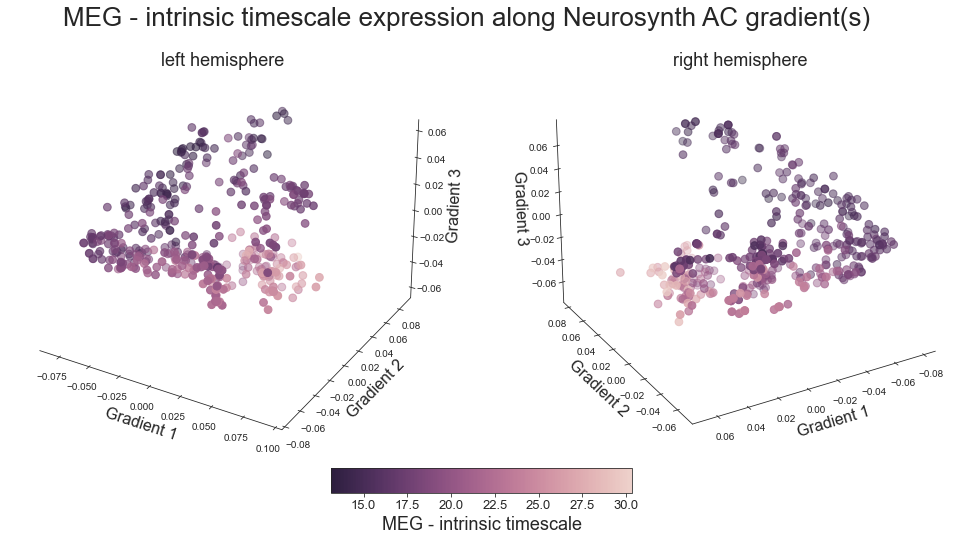
function#
rs-fMRI Intra-subject variability#
mueller_intersubjvar_maps = datasets.fetch_annotation(source='mueller2013', desc='intersubjvar')
mueller_intersubjvar_map_left_fsaverage = transforms.fslr_to_fsaverage(mueller_intersubjvar_maps[0], '10k', hemi='L')[0]
mueller_intersubjvar_map_right_fsaverage = transforms.fslr_to_fsaverage(mueller_intersubjvar_maps[1], '10k', hemi='R')[0]
plot_surf_template((mueller_intersubjvar_map_left_fsaverage, mueller_intersubjvar_map_right_fsaverage), 'fsaverage', '10k', cmap=cmr.eclipse);

mueller_intersubjvar_map_left_fsaverage_mask_ac, mueller_intersubjvar_map_left_fsaverage_mask_ac_data = mask_surface(mueller_intersubjvar_map_left_fsaverage, ac_mask_surface[0])
mueller_intersubjvar_map_right_fsaverage_mask_ac, mueller_intersubjvar_map_right_fsaverage_mask_ac_data = mask_surface(mueller_intersubjvar_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((mueller_intersubjvar_map_left_fsaverage_mask_ac, mueller_intersubjvar_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmr.eclipse);
compute_variograms(mueller_intersubjvar_map_left_fsaverage_mask_ac_data, mueller_intersubjvar_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
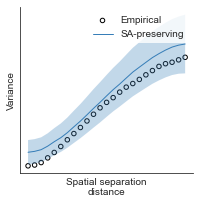
Computing variogram for right hemisphere
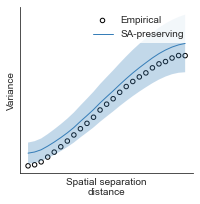
mueller_intersubjvar_map_left_fsaverage_mask_ac_surrogates, mueller_intersubjvar_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(mueller_intersubjvar_map_left_fsaverage_mask_ac_data,
mueller_intersubjvar_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(mueller_intersubjvar_map_left_fsaverage_mask_ac_surrogates,
mueller_intersubjvar_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmr.eclipse)
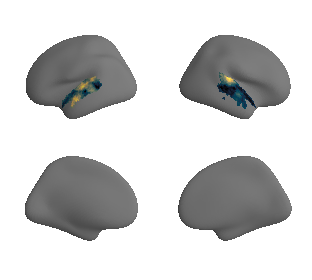

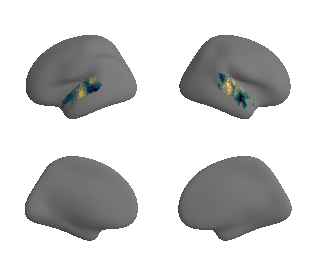

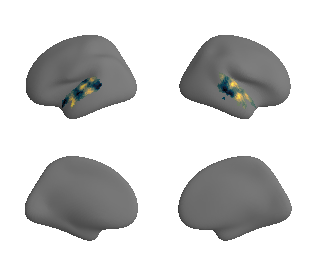
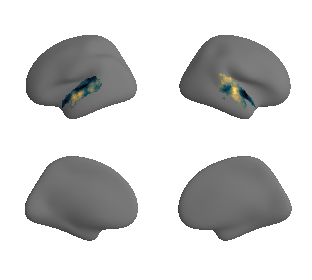


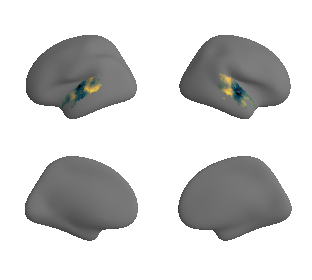
grad_intersubjvar_cors = compare_brain_maps_surrogates(mueller_intersubjvar_map_left_fsaverage_mask_ac_data, mueller_intersubjvar_map_right_fsaverage_mask_ac_data,
mueller_intersubjvar_map_left_fsaverage_mask_ac_surrogates, mueller_intersubjvar_map_right_fsaverage_mask_ac_surrogates,
feature='Inter-subject variability',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_intersubjvar_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Inter-subject variability are correlated with an r of 0.6413602134517691 at p = 0.015
Working on gradient 2
The LH maps of gradient 2 & Inter-subject variability are correlated with an r of 0.25894381150001555 at p = 0.454
Working on gradient 3
The LH maps of gradient 3 & Inter-subject variability are correlated with an r of -0.07461041002677535 at p = 0.85
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Inter-subject variability are correlated with an r of 0.669794576193322 at p = 0.028
Working on gradient 2
The RH maps of gradient 2 & Inter-subject variability are correlated with an r of 0.27449300061301785 at p = 0.373
Working on gradient 3
The RH maps of gradient 3 & Inter-subject variability are correlated with an r of -0.13848697665717333 at p = 0.73
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_intersubjvar_cors.json
plot_3d_gradients(mueller_intersubjvar_map_left_fsaverage, mueller_intersubjvar_map_right_fsaverage, 'rs-fMRI inter-subject variability', 'rs-fMRI inter-subject variability', cmap=cmr.eclipse)
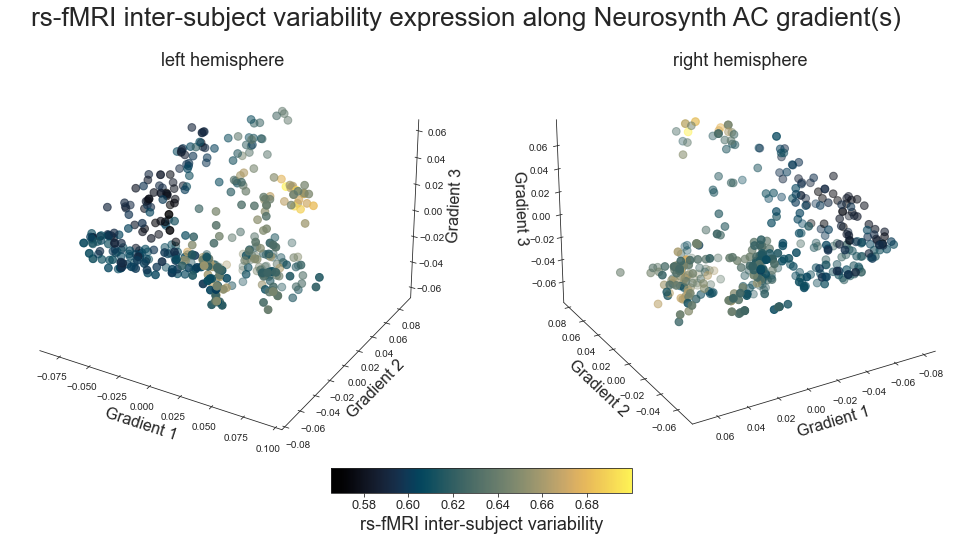
FC 1st gradient#
margulies_gradient1_maps = datasets.fetch_annotation(source='margulies2016', desc='fcgradient01')
margulies_gradient1_map_left_fsaverage = transforms.fslr_to_fsaverage(margulies_gradient1_maps[0], '10k', hemi='L')[0]
margulies_gradient1_map_right_fsaverage = transforms.fslr_to_fsaverage(margulies_gradient1_maps[1], '10k', hemi='R')[0]
plot_surf_template((margulies_gradient1_map_left_fsaverage, margulies_gradient1_map_right_fsaverage), 'fsaverage', '10k', cmap=cmr.eclipse);
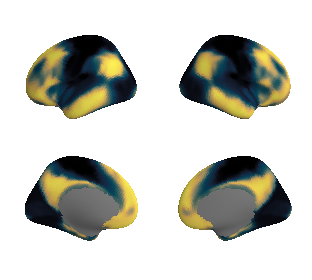
margulies_gradient1_map_left_fsaverage_mask_ac, margulies_gradient1_map_left_fsaverage_mask_ac_data = mask_surface(margulies_gradient1_map_left_fsaverage, ac_mask_surface[0])
margulies_gradient1_map_right_fsaverage_mask_ac, margulies_gradient1_map_right_fsaverage_mask_ac_data = mask_surface(margulies_gradient1_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((margulies_gradient1_map_left_fsaverage_mask_ac, margulies_gradient1_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmr.eclipse);

compute_variograms(margulies_gradient1_map_left_fsaverage_mask_ac_data, margulies_gradient1_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
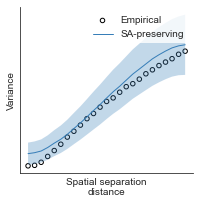
Computing variogram for right hemisphere
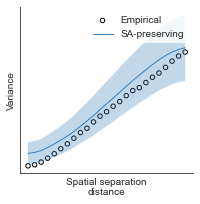
margulies_gradient1_map_left_fsaverage_mask_ac_surrogates, margulies_gradient1_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(margulies_gradient1_map_left_fsaverage_mask_ac_data,
margulies_gradient1_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(margulies_gradient1_map_left_fsaverage_mask_ac_surrogates,
margulies_gradient1_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmr.eclipse)
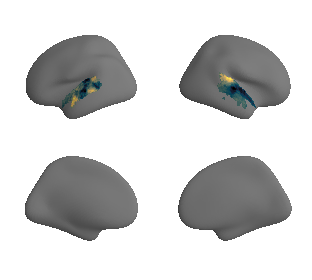
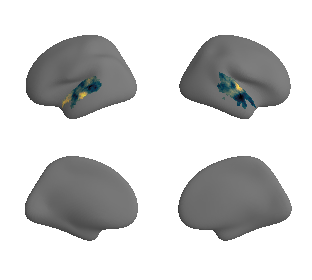



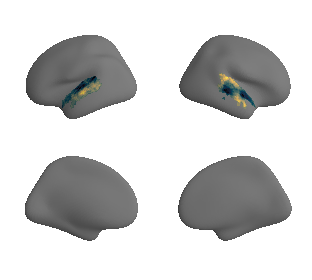


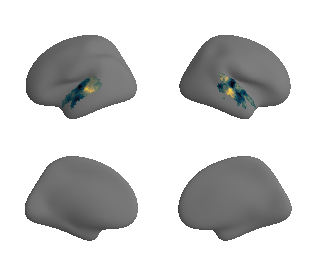
grad_margulies_gradient_1_cors = compare_brain_maps_surrogates(margulies_gradient1_map_left_fsaverage_mask_ac_data, margulies_gradient1_map_right_fsaverage_mask_ac_data,
margulies_gradient1_map_left_fsaverage_mask_ac_surrogates, margulies_gradient1_map_right_fsaverage_mask_ac_surrogates,
feature='FC Gradient 1',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_margulies_gradient_1_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & FC Gradient 1 are correlated with an r of 0.6576739908212915 at p = 0.013
Working on gradient 2
The LH maps of gradient 2 & FC Gradient 1 are correlated with an r of 0.0074710662848029175 at p = 0.985
Working on gradient 3
The LH maps of gradient 3 & FC Gradient 1 are correlated with an r of 0.028351848248585165 at p = 0.939
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & FC Gradient 1 are correlated with an r of 0.6918935242405332 at p = 0.027
Working on gradient 2
The RH maps of gradient 2 & FC Gradient 1 are correlated with an r of 0.0028252917192951865 at p = 0.994
Working on gradient 3
The RH maps of gradient 3 & FC Gradient 1 are correlated with an r of -0.13372388388485545 at p = 0.782
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_margulies_gradient_1_cors.json
plot_3d_gradients(margulies_gradient1_map_left_fsaverage, margulies_gradient1_map_right_fsaverage, 'FC gradient 1', 'FC gradient 1', cmap=cmr.eclipse)
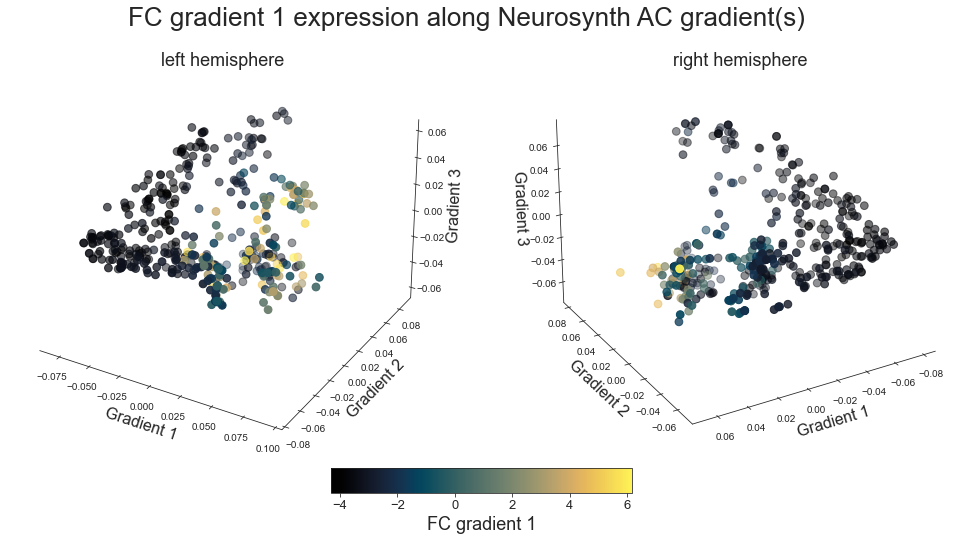
Sensory-association mean rank axis#
sydnor_saaxis_maps = datasets.fetch_annotation(source='sydnor2021', desc='SAaxis')
sydnor_saaxis_map_left_fsaverage = transforms.fslr_to_fsaverage(sydnor_saaxis_maps[0], '10k', hemi='L')[0]
sydnor_saaxis_map_right_fsaverage = transforms.fslr_to_fsaverage(sydnor_saaxis_maps[1], '10k', hemi='R')[0]
plot_surf_template((sydnor_saaxis_map_left_fsaverage, sydnor_saaxis_map_right_fsaverage), 'fsaverage', '10k', cmap=cmr.eclipse);
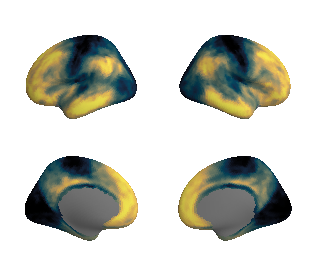
sydnor_saaxis_map_left_fsaverage_mask_ac, sydnor_saaxis_map_left_fsaverage_mask_ac_data = mask_surface(sydnor_saaxis_map_left_fsaverage, ac_mask_surface[0])
sydnor_saaxis_map_right_fsaverage_mask_ac, sydnor_saaxis_map_right_fsaverage_mask_ac_data = mask_surface(sydnor_saaxis_map_right_fsaverage, ac_mask_surface[1])
plot_surf_template((sydnor_saaxis_map_left_fsaverage_mask_ac, sydnor_saaxis_map_right_fsaverage_mask_ac), 'fsaverage', '10k', cmap=cmr.eclipse);

compute_variograms(sydnor_saaxis_map_left_fsaverage_mask_ac_data, sydnor_saaxis_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
Computing variogram for left hemisphere
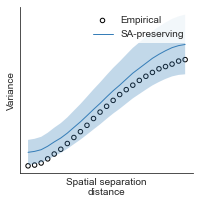
Computing variogram for right hemisphere
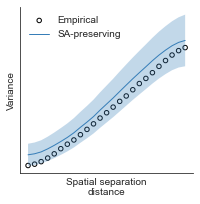
sydnor_saaxis_map_left_fsaverage_mask_ac_surrogates, sydnor_saaxis_map_right_fsaverage_mask_ac_surrogates = compute_surrogate_maps(sydnor_saaxis_map_left_fsaverage_mask_ac_data,
sydnor_saaxis_map_right_fsaverage_mask_ac_data,
ns=50, knn=200)
working on LH surrogates
working on RH surrogates
plot_surrogate_maps(sydnor_saaxis_map_left_fsaverage_mask_ac_surrogates,
sydnor_saaxis_map_right_fsaverage_mask_ac_surrogates, n_sur=10, cmap=cmr.eclipse)
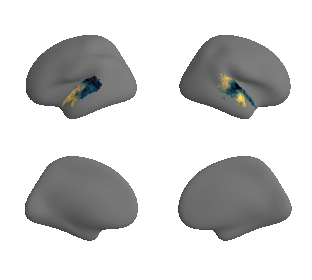

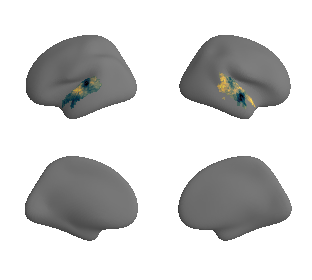
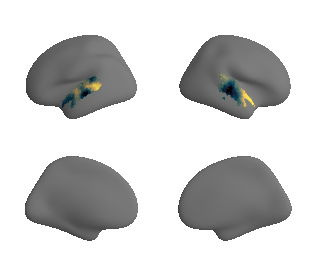

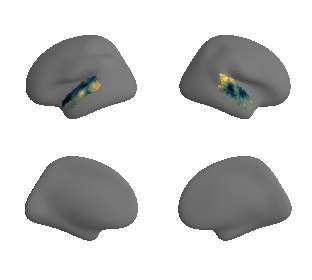
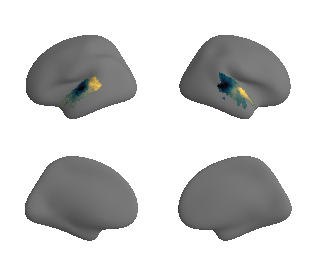

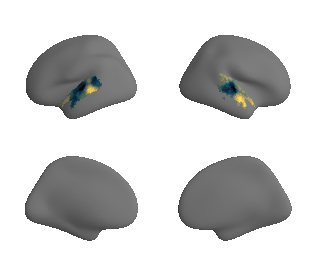
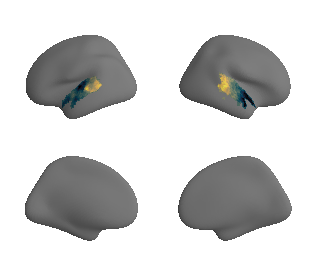
grad_saaxis_cors = compare_brain_maps_surrogates(sydnor_saaxis_map_left_fsaverage_mask_ac_data, sydnor_saaxis_map_right_fsaverage_mask_ac_data,
sydnor_saaxis_map_left_fsaverage_mask_ac_surrogates, sydnor_saaxis_map_right_fsaverage_mask_ac_surrogates,
feature='Sensory-association mean rank',
path='/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_saaxis_cors.json')
Working on LH maps
Working on gradient 1
The LH maps of gradient 1 & Sensory-association mean rank are correlated with an r of 0.7418539385996007 at p = 0.002
Working on gradient 2
The LH maps of gradient 2 & Sensory-association mean rank are correlated with an r of 0.29976055123781953 at p = 0.406
Working on gradient 3
The LH maps of gradient 3 & Sensory-association mean rank are correlated with an r of -0.25917274353313163 at p = 0.514
Working on RH maps
Working on gradient 1
The RH maps of gradient 1 & Sensory-association mean rank are correlated with an r of 0.796229405298785 at p = 0.003
Working on gradient 2
The RH maps of gradient 2 & Sensory-association mean rank are correlated with an r of 0.1698912234071549 at p = 0.644
Working on gradient 3
The RH maps of gradient 3 & Sensory-association mean rank are correlated with an r of -0.3367478829937502 at p = 0.497
The results will be save to /Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_saaxis_cors.json
plot_3d_gradients(sydnor_saaxis_map_left_fsaverage, sydnor_saaxis_map_right_fsaverage, 'Sensory-association mean rank', 'Sensory-association mean rank', cmap=cmr.eclipse)
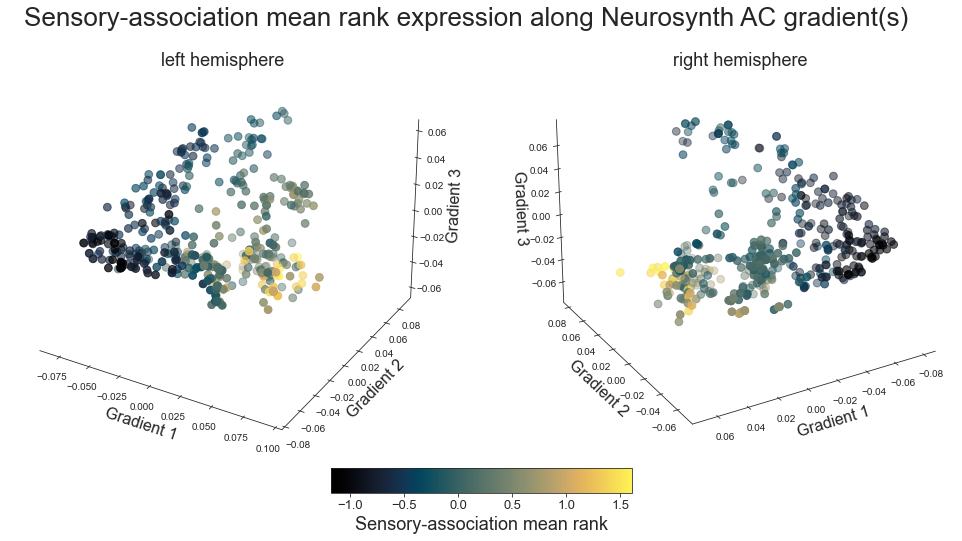
Gather all outcomes in one DataFrame#
list_cor_dicts = [grad_hcp_myelin_cors, grad_hcp_thickness_cors, grad_genepc1_cors,
grad_cbf_cors, grad_oxy_cors, grad_glu_cors,
grad_evoexp_cors, grad_fchomology_cors, grad_scaling_cors,
grad_meg_alpha_cors, grad_meg_beta_cors, grad_meg_delta_cors, grad_meg_theta_cors, grad_meg_timescale_cors,
grad_intersubjvar_cors, grad_margulies_gradient_1_cors, grad_saaxis_cors]
list_cors = [cor_dict[hemi][grad]['r'] for grad in ['grad_1', 'grad_2', 'grad_3'] for hemi in ['LH', 'RH'] for cor_dict in list_cor_dicts]
list_cor_ps = [cor_dict[hemi][grad]['p'] for grad in ['grad_1', 'grad_2', 'grad_3'] for hemi in ['LH', 'RH'] for cor_dict in list_cor_dicts]
list_cors_perm = [cor_dict[hemi][grad]['cors'] for grad in ['grad_1', 'grad_2', 'grad_3'] for hemi in ['LH', 'RH'] for cor_dict in list_cor_dicts]
list_features = ['myelin', 'thickness', 'abagen', 'CBF',
'oxygen', 'glucose', 'evolutionary expansion', 'cross-species FH',
'cortical areal scaling', 'MEG-Alpha', 'MEG-Beta', 'MEG-Delta',
'MEG-Theta', 'intrinsic timescale', 'rs-fMRI intersubjvar',
'FC gradient-1', 'sensory-association axis']
list_features_nest = [[feature] *6 for feature in list_features]
list_features_all = [feat for feature in list_features_nest for feat in feature]
list_gradients = ['gradient_1', 'gradient_2', 'gradient_3']
grad_feature_cor_df = pd.DataFrame({'feature_map': list_features_all, 'gradient': list_gradients*(len(list_features)*2),
'hemisphere': ['LH', 'RH'] * (len(list_features)*3), 'correlation': list_cors,
'p value': list_cor_ps, 'correlation permuted': list_cors_perm})
grad_feature_cor_df
| feature_map | gradient | hemisphere | correlation | p value | correlation permuted | |
|---|---|---|---|---|---|---|
| 0 | myelin | gradient_1 | LH | -0.765486 | 0.014 | [-0.22844491498226122, -0.7790571643127192, 0.... |
| 1 | myelin | gradient_2 | RH | 0.447029 | 0.171 | [0.33651523175223597, 0.5229881213025092, -0.0... |
| 2 | myelin | gradient_3 | LH | -0.681142 | 0.026 | [0.34426836695905494, -0.7501520620890701, 0.2... |
| 3 | myelin | gradient_1 | RH | -0.748699 | 0.005 | [-0.3049778011268198, -0.7276647597567475, 0.4... |
| 4 | myelin | gradient_2 | LH | -0.571523 | 0.085 | [0.7165362703629115, -0.6041640168819621, 0.63... |
| ... | ... | ... | ... | ... | ... | ... |
| 97 | sensory-association axis | gradient_2 | RH | 0.069287 | 0.870 | [-0.28665592458548633, 0.7522813208844119, 0.3... |
| 98 | sensory-association axis | gradient_3 | LH | -0.548593 | 0.243 | [0.2589649224531136, -0.08635056383521521, 0.2... |
| 99 | sensory-association axis | gradient_1 | RH | -0.138487 | 0.730 | [0.4399991982804856, 0.1060072907296043, 0.099... |
| 100 | sensory-association axis | gradient_2 | LH | -0.133724 | 0.782 | [0.6012462160673157, 0.08232403441262304, 0.14... |
| 101 | sensory-association axis | gradient_3 | RH | -0.336748 | 0.497 | [0.3431025598542449, -0.8056972534383884, 0.26... |
102 rows × 6 columns
grad_feature_cor_df.to_csv('/Users/peerherholz/google_drive/GitHub/ns_ac_walkthrough/walkthrough/results/gradients/gradient_profiling/grad_feature_cor_all.csv')