Research
Comparing generalization in biological & artificial networks
Building upon my research in auditory processing, I investigate generalization in biological and artificial agents and the potentially underlying transformations as a basis for intelligent behavior. More specifically, I’m interested in the computations and resulting transformations agents and systems conduct from incoming sensory information along a processing cascade to achieve abstract representations that allow generalization across contexts and modalities. Within that I furthermore focus on the how these processes can be compared between biological and artificial networks, as well as how the first can inform the latter to achieve a better performance. Here, I utilize fMRI and ANNs on a broad range of tasks, including “classic” and but especially naturalistic paradigms.
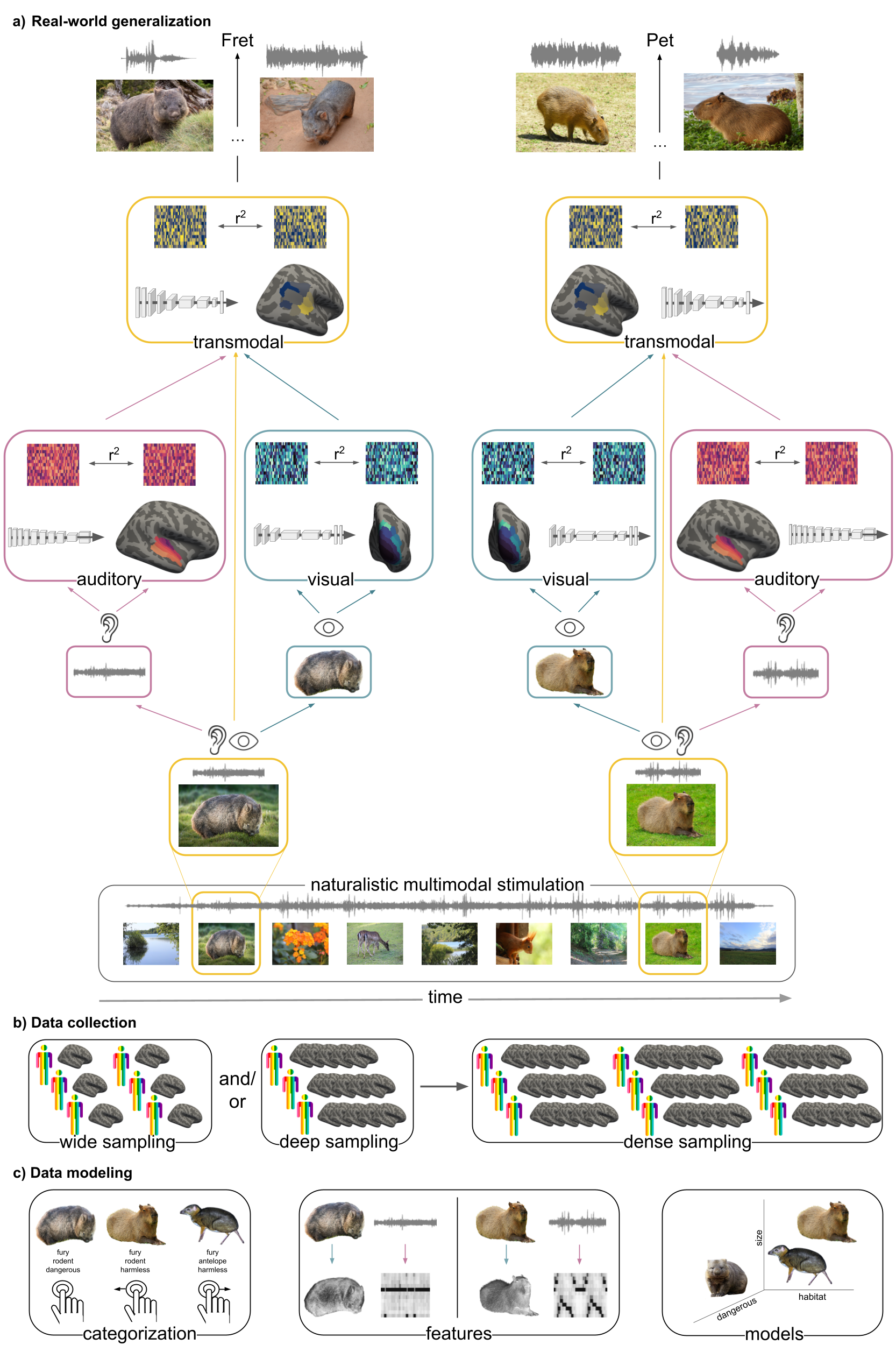
The auditory system as a model mechanism of complex behavior
The auditory system remains a complex and evolving field despite advancements in data acquisition and analysis. I explore its functional organization, including structure-function relationships, diverging pathways, and communication between cortical and subcortical areas using ultra-high field MRI. Furthermore, my research investigates how various sound categories, including music, language, and singing, are processed and differentiated in these pathways, utilizing a combination of MRI, EEG, and machine learning. Factors like musical training and development are also considered in studying music perception, aiming to understand and describe how different brains perceive and categorize music through a variety of methods including imaging, behavioral studies, and computational models.
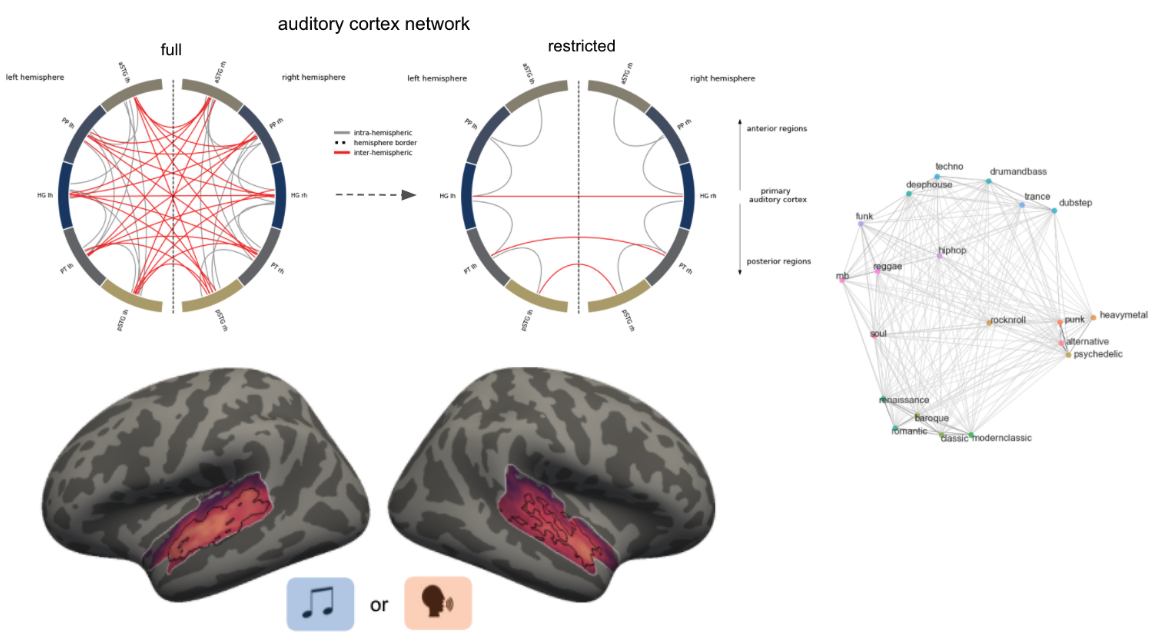
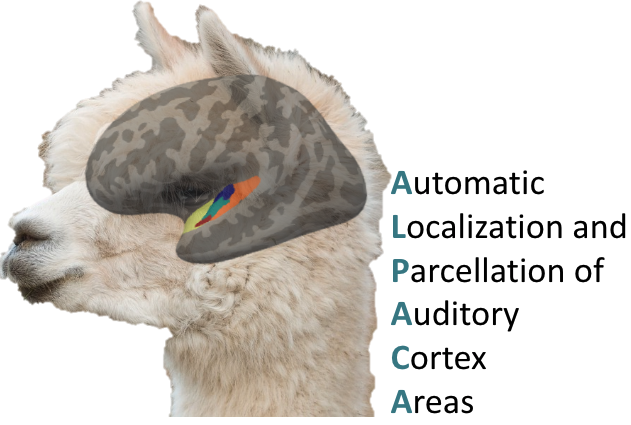
Tools for auditory neuroscience
Auditory neuroscience research, particularly with MRI, faces unique challenges such as complex stimulus experiments, prolonged durations, low signal-to-noise ratios and unreliable localization and parcellation of auditory cortex areas. Addressing these issues, I work on enhancing experimental settings, including optimizing MRI acquisition parameters like ISSS and multiband, and developing software packages. For example, ANSL, that allows audiometry measurements in the MRI environment and ALPACA, including experiment and analyses scripts for different paradigms (natural sounds & classic tone bursts) and analysis approaches (structural parcellation, mapping of auditory ROIs from atlases, fMRI, EEG, searchlights, encoding, etc.) to locate and parcellate auditory cortex areas. I also worked in an online repository where folks can upload the respective outputs of these software packages along with meta-data like field strength and analysis approach that will allow comprehensive meta-analyses and exploration of results from previous research work.
Neuroinformatics & methods
I was once told that one doesn’t have to understand neuroscience methods and statistics in order to apply them. While this is certainly true, it’s also certainly bad. Starting with very little method & zero programming skills everything was kinda overwhelming but at the same time fascinating. The combination of math, physics, informatics and biology amazed me and the more I read and gained practical experience the more I wanted to understand. I’m especially interested in data management and quality control, non-standard experimental setups (e.g., naturalistic stimulation) and acquisition schemes (e.g., multiband, MT), processing steps (e.g., ICA, detrending), image registration, multi-modal measurements & data integration, as well as multivariate analyses approaches (e.g., machine learning, encoding models). Furthermore, I’m working on pipeline creation / automation and cloud / hpc computing and setting up server systems.

Open & reproducible (neuro)science
Starting with the lack of details in the methods section of papers I was reading to no chance of having a look at the data or code (or sometimes even the whole publication) created some sort of frustration I found it hard to cope with. Hence, I’m trying to open up my daily research workflow as much as possible throughout all stages using a variety of tools. Through the support during my time as an “open science fellow” I was able to initiate the Open Science Initiative University of Marburg, an university wide organization fostering open science principles for example via hackrooms and hackathons (e.g. brainhacks), workshops, as well as general assistance and support. Additionally, I also became part of the BIDS and ReproNim teams to foster data standardization, provenance tracking and reproducibility, as well as generalizability.