Introduction to SciPy
Contents
Introduction to SciPy¶
Disclaimer: Most of the content in this notebook is coming from www.scipy-lectures.org
Scipy - High-level Scientific Computing¶
The scipy package contains various toolboxes dedicated to common issues in scientific computing. Its different submodules correspond to different applications, such as interpolation, integration, optimization, image processing, statistics, special functions, etc.
scipy is composed of task-specific sub-modules:
scipy.cluster Clustering package
scipy.constants Constants
scipy.fftpack Discrete Fourier transforms
scipy.integrate Integration and ODEs
scipy.interpolate Interpolation
scipy.io Input and output
scipy.linalg Linear algebra
scipy.misc Miscellaneous routines
scipy.ndimage Multi-dimensional image processing
scipy.odr Orthogonal distance regression
scipy.optimize Optimization and root finding
scipy.signal Signal processing
scipy.sparse Sparse matrices
scipy.sparse.linalg Sparse linear algebra
scipy.sparse.csgraph Compressed Sparse Graph Routines
scipy.spatial Spatial algorithms and data structures
scipy.special Special functions
scipy.stats Statistical functions
scipy.stats.mstats Statistical functions for masked arrays
They all depend on numpy, but are mostly independent of each
other. The standard way of importing Numpy and these Scipy modules
is:
import numpy as np
from scipy import stats # same for other sub-modules
Warning: This tutorial is far from a complete introduction to numerical computing. As enumerating the different submodules and functions in scipy would be very boring, we concentrate instead on a few examples to give a general idea of how to use scipy for scientific computing.
File input/output: scipy.io¶
Load and Save MATLAB files¶
from scipy import io as spio
a = np.ones((3, 3))
spio.savemat('file.mat', {'a': a}) # savemat expects a dictionary
ls *mat
file.mat
data = spio.loadmat('file.mat')
data['a']
array([[1., 1., 1.],
[1., 1., 1.],
[1., 1., 1.]])
See also¶
Load text files:
numpy.loadtxt/numpy.savetxtClever loading of text/csv files:
numpy.genfromtxt/numpy.recfromcsvFast and efficient, but numpy-specific, binary format:
numpy.save/numpy.loadMore advanced input/output of images in scikit-image:
skimage.io
Linear algebra operations: scipy.linalg¶
The scipy.linalg module provides standard linear algebra operations.
The scipy.linalg.det function computes the determinant of a square matrix:
from scipy import linalg
arr = np.array([[1, 2],
[3, 4]])
linalg.det(arr)
-2.0
The scipy.linalg.inv function computes the inverse of a square matrix:
arr = np.array([[1, 2],
[3, 4]])
iarr = linalg.inv(arr)
iarr
array([[-2. , 1. ],
[ 1.5, -0.5]])
More advanced operations are available, for example singular-value decomposition (SVD):
arr = np.arange(9).reshape((3, 3)) + np.diag([1, 0, 1])
arr
array([[1, 1, 2],
[3, 4, 5],
[6, 7, 9]])
uarr, spec, vharr = linalg.svd(arr)
# The resulting array spectrum is
spec
array([14.88982544, 0.45294236, 0.29654967])
The original matrix can be re-composed by matrix multiplication of the outputs of svd with np.dot:
sarr = np.diag(spec)
svd_mat = uarr.dot(sarr).dot(vharr)
svd_mat
array([[1., 1., 2.],
[3., 4., 5.],
[6., 7., 9.]])
SVD is commonly used in statistics and signal processing. Many other standard decompositions (QR, LU, Cholesky, Schur), as well as solvers for linear systems, are available in scipy.linalg.
Interpolation: scipy.interpolate¶
scipy.interpolate is useful for fitting a function from experimental data and thus evaluating points where no measure exists.
# Let's create some data
measured_time = np.linspace(0, 1, 10)
noise = (np.random.random(10)*2 - 1) * 1e-1
measures = np.sin(2 * np.pi * measured_time) + noise
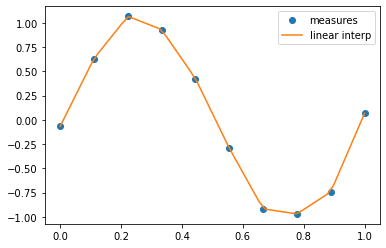
scipy.interpolate.interp1d can build a linear interpolation
function:
from scipy.interpolate import interp1d
linear_interp = interp1d(measured_time, measures)
interpolation_time = np.linspace(0, 1, 50)
linear_results = linear_interp(interpolation_time)
%matplotlib inline
from matplotlib import pyplot as plt
plt.plot(measured_time, measures, 'o', ms=6, label='measures')
plt.plot(interpolation_time, linear_results, label='linear interp')
plt.legend();
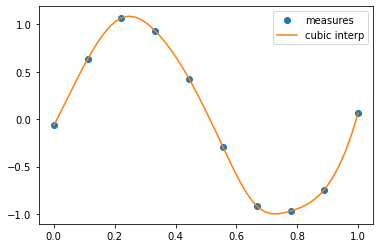
A cubic interpolation can also be selected by providing the kind optional keyword argument:
cubic_interp = interp1d(measured_time, measures, kind='cubic')
cubic_results = cubic_interp(interpolation_time)
plt.plot(measured_time, measures, 'o', ms=6, label='measures')
plt.plot(interpolation_time, cubic_results, label='cubic interp')
plt.legend()
<matplotlib.legend.Legend at 0x7f1e0bbe0fd0>
Optimization and fit: scipy.optimize¶
Optimization is the problem of finding a numerical solution to a minimization or equality.
The scipy.optimize module provides algorithms for function minimization (scalar or multi-dimensional), curve fitting and root finding.
from scipy import optimize
Curve fitting¶
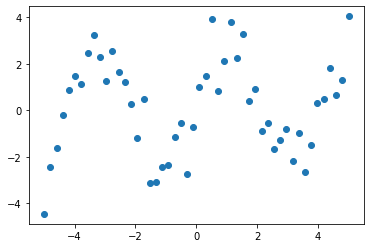
Suppose we have data on a sine wave, with some noise:
x_data = np.linspace(-5, 5, num=50)
y_data = 2.9 * np.sin(1.5 * x_data) + np.random.normal(size=50)
# And plot it
import matplotlib.pyplot as plt
plt.figure(figsize=(6, 4))
plt.scatter(x_data, y_data)
plt.show()
If we know that the data lies on a sine wave, but not the amplitudes or the period, we can find those by least squares curve fitting. First, we have to define the test function to fit, here a sine with unknown amplitude and period:
def test_func(x, a, b):
return a * np.sin(b * x)
We then use scipy.optimize.curve_fit to find \(a\) and \(b\):
params, params_covariance = optimize.curve_fit(
test_func, x_data, y_data, p0=[2, 2])
print(params)
[2.58100495 1.50430612]
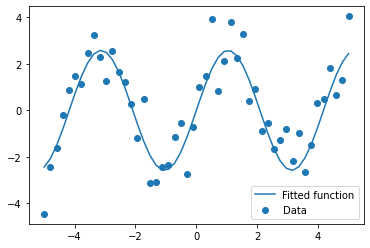
# And plot the resulting curve on the data
plt.figure(figsize=(6, 4))
plt.scatter(x_data, y_data, label='Data')
plt.plot(x_data, test_func(x_data, params[0], params[1]),
label='Fitted function')
plt.legend(loc='best')
plt.show()
Finding the minimum of a scalar function¶
Let’s define the following function:
def f(x):
return x**2 + 10*np.sin(x)
and plot it:
x = np.arange(-10, 10, 0.1)
plt.plot(x, f(x))
plt.show()
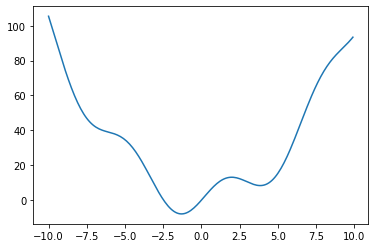
This function has a global minimum around -1.3 and a local minimum around 3.8.
Searching for minimum can be done with scipy.optimize.minimize, given a starting point x0, it returns the location of the minimum that it has found:
result = optimize.minimize(f, x0=0)
result
fun: -7.945823375615215
hess_inv: array([[0.08589237]])
jac: array([-1.1920929e-06])
message: 'Optimization terminated successfully.'
nfev: 18
nit: 5
njev: 6
status: 0
success: True
x: array([-1.30644012])
result.x # The coordinate of the minimum
array([-1.30644012])
Fast Fourier transforms: scipy.fftpack¶
The scipy.fftpack module computes fast Fourier transforms (FFTs)
and offers utilities to handle them. The main functions are:
scipy.fftpack.fftto compute the FFTscipy.fftpack.fftfreqto generate the sampling frequenciesscipy.fftpack.ifftcomputes the inverse FFT, from frequency space to signal space
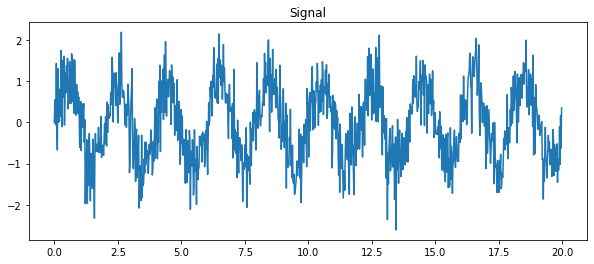
As an illustration, let’s create a (noisy) input signal (sig), and its FFT:
from scipy import fftpack
time_step = 0.02
period = 1. / 0.5 # 0.5 Hz
time_vec = np.arange(0, 20, time_step)
sig = (np.sin(2 * np.pi / period * time_vec)
+ 0.5 * np.random.randn(time_vec.size))
plt.figure(figsize=(10, 4))
plt.plot(time_vec, sig, label='Original signal')
plt.title('Signal')
plt.show()
# The FFT of the signal
sig_fft = fftpack.fft(sig)
# And the power (sig_fft is of complex dtype)
power = np.abs(sig_fft)
# The corresponding frequencies
sample_freq = fftpack.fftfreq(sig.size, d=time_step)
# Plot the FFT power
plt.figure(figsize=(10, 4))
plt.plot(sample_freq[:500], power[:500])
plt.xlabel('Frequency [Hz]')
plt.ylabel('plower')
plt.title('FFT')
plt.show()
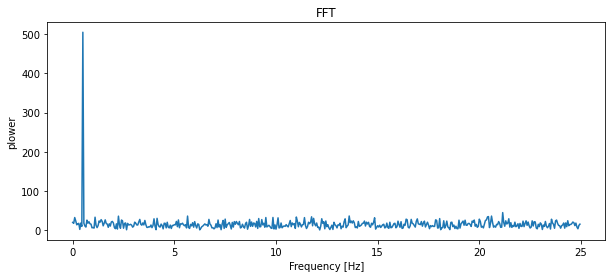
The peak signal frequency can be found with sample_freq[power.argmax()]
peak_freq = sample_freq[power.argmax()]
peak_freq
0.5
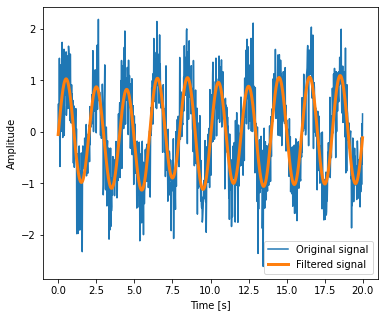
Setting the Fourier component above this frequency to zero and inverting the FFT with scipy.fftpack.ifft, gives a filtered signal:
high_freq_fft = sig_fft.copy()
high_freq_fft[np.abs(sample_freq) > peak_freq] = 0
filtered_sig = fftpack.ifft(high_freq_fft)
plt.figure(figsize=(6, 5))
plt.plot(time_vec, sig, label='Original signal')
plt.plot(time_vec, filtered_sig, linewidth=3, label='Filtered signal')
plt.xlabel('Time [s]')
plt.ylabel('Amplitude')
plt.legend(loc='best')
plt.show();
/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/numpy/core/_asarray.py:85: ComplexWarning: Casting complex values to real discards the imaginary part
return array(a, dtype, copy=False, order=order)
Signal processing: scipy.signal¶
scipy.signal is for typical signal processing: 1D, regularly-sampled signals.
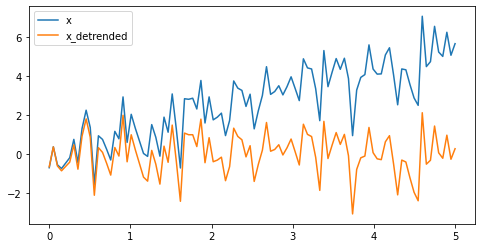
Detrending scipy.signal.detrend - remove linear trend from signal:¶
# Generate a random signal with a trend
import numpy as np
t = np.linspace(0, 5, 100)
x = t + np.random.normal(size=100)
# Detrend
from scipy import signal
x_detrended = signal.detrend(x)
# Plot
from matplotlib import pyplot as plt
plt.figure(figsize=(8, 4))
plt.plot(t, x, label="x")
plt.plot(t, x_detrended, label="x_detrended")
plt.legend(loc='best')
plt.show()