Data analyzes II - data visualization and analyses
Contents
Data analyzes II - data visualization and analyses¶
Peer Herholz (he/him)
Habilitation candidate - Fiebach Lab, Neurocognitive Psychology at Goethe-University Frankfurt
Research affiliate - NeuroDataScience lab at MNI/McGill
Member - BIDS, ReproNim, Brainhack, Neuromod, OHBM SEA-SIG, UNIQUE
![]()
![]() @peerherholz
@peerherholz
Before we get started …¶
most of what you’ll see within this lecture was prepared by Ross Markello, Michael Notter and Peer Herholz and further adapted for this course by Peer Herholz
based on Tal Yarkoni’s “Introduction to Python” lecture at Neurohackademy 2019
based on 10 minutes to pandas
Objectives 📍¶
learn basic and efficient usage of python for data analyzes & visualization
working with data:
reading, working, writing
preprocessing, filtering, wrangling
visualizing data:
basic plots
advanced & fancy stuff
analyzing data:
descriptive stats
inferential stats
Recap - Why do data science in Python?¶
all the general benefits of the
Python language(open source, fast, etc.)Specifically, it’s a widely used/very flexible, high-level, general-purpose, dynamic programming language
the
Python ecosystemcontains tens of thousands of packages, several are very widely used in data science applications:Jupyter: interactive notebooks
Numpy: numerical computing in Python
pandas: data structures for Python
pingouin: statistics in Python
statsmodels: statistics in Python
seaborn: data visualization in Python
plotly: interactive data visualization in Python
Scipy: scientific Python tools
Matplotlib: plotting in Python
scikit-learn: machine learning in Python
even more:
Pythonhas very good (often best-in-class) external packages for almost everythingParticularly important for data science, which draws on a very broad toolkit
Package management is easy (conda, pip)
Examples for further important/central python data science packages :
Web development: flask, Django
Database ORMs: SQLAlchemy, Django ORM (w/ adapters for all major DBs)
Scraping/parsing text/markup: beautifulsoup, scrapy
Natural language processing (NLP): nltk, gensim, textblob
Numerical computation and data analysis: numpy, scipy, pandas, xarray
Machine learning: scikit-learn, Tensorflow, keras
Image processing: pillow, scikit-image, OpenCV
Plotting: matplotlib, seaborn, altair, ggplot, Bokeh
GUI development: pyQT, wxPython
Testing: py.test
Widely-used¶
Python is the fastest-growing major programming language
Top 3 overall (with JavaScript, Java)

What we will do in this section of the course is a short introduction to Python for data analyses including basic data operations like file reading and wrangling, as well as statistics and data visualization. The goal is to showcase crucial tools/resources and their underlying working principles to allow further more in-depth exploration and direct application.
It is divided into the following chapters:
Getting ready
Basic data operations
Reading data
Exploring data
Data wrangling
Basic data visualization
Underlying principles
“standard” plots
Going further with advanced plots
Statistics in python
Descriptive analyses
Inferential analyses
Interactive data visualization
Here’s what we will focus on in the second block:
Basic data visualization
Underlying principles
“standard” plots
Going further with advanced plots
Statistics in python
Descriptive analyses
Inferential analyses
Interactive data visualization
Recap - Getting ready¶
What’s the first thing we have to check/evaluate before we start working with data, no matter if in Python or any other software? That’s right: getting everything ready!
This includes outlining the core workflow and respective steps. Quite often, this notebook and its content included, this entails the following:
What kind of data do I have and where is it?
What is the goal of the data analyses?
How will the respective steps be implemented?
So let’s check these aspects out in slightly more detail.
Recap - What kind of data do I have and where is it¶
The first crucial step is to get a brief idea of the kind of data we have, where it is, etc. to outline the subsequent parts of the workflow (python modules to use, analyses to conduct, etc.). At this point it’s important to note that Python and its modules work tremendously well for basically all kinds of data out there, no matter if behavior, neuroimaging, etc. . To keep things rather simple, we will keep using the behavioral dataset from last session that contains ratings and demographic information from a group of university students (ah, the classics…).
We already accomplished and worked with the dataset quite a bit during the last session, including:
reading data
extract data of interest
convert to different more intelligible structures and forms
At the end, we had two dataframes, both containing the data of all participants but in different formats. Does anyone remember what formats datasets commonly have and how they differ?
True that, we initially had a dataframe in wide-format but then created a reshaped version in long-format. For this session, we will continue to explore aspects of data visualization and analyzes via this dataframe. Thus, let’s move to our data storage and analyses directory and load it accordingly using pandas!
from os import chdir
chdir('/Users/peerherholz/Desktop/data_experiment/')
from os import listdir
listdir('.')
['yagurllea_experiment_2022_Jan_25_1318.csv',
'pauli4psychopy_experiment_2022_Jan_24_1145.csv',
'porla_experiment_2022_Jan_24_2154.csv',
'Nina__experiment_2022_Jan_24_1212.csv',
'.DS_Store',
'derivatives',
'participant_123_experiment_2022_Jan_26_1307.csv',
'funny_ID_experiment_2022_Jan_24_1215.csv',
'Smitty_Werben_Jagger_Man_Jensen#1_experiment_2022_Jan_26_1158.csv',
'wile_coyote_2022_Jan_23_1805.csv',
'Sophie_Oprée_experiment_2022_Jan_24_1411.csv',
'Heidi_Klum_experiment_2022_Jan_26_1600.csv',
'bugs_bunny_experiment_2022_Jan_23_1745.csv',
'Bruce_Wayne_experiment_2022_Jan_23_1605.csv',
'twentyfour_experiment_2022_Jan_26_1436.csv',
'Chiara_Ferrandina_experiment_2022_Jan_24_1829.csv',
'daffy_duck_experiment_2022_Jan_23_1409.csv']
listdir('derivatives/preprocessing/')
['Smitty_Werben_Jagger_Man_Jensen#1_long_format.csv',
'dataframe_group.csv',
'bugs_bunny_long_format.csv',
'pauli4psychopy_long_format.csv',
'Heidi Klum_long_format.csv',
'Sophie Oprée_long_format.csv',
'daffy_duck_long_format.csv',
'Nina _long_format.csv',
'wile_coyote_long_format.csv',
'funny ID_long_format.csv',
'yagurllea_long_format.csv',
'participant_123_long_format.csv',
'porla_long_format.csv',
'Chiara Ferrandina_long_format.csv',
'Bruce_Wayne_long_format.csv',
'Talha_long_format.csv']
import pandas as pd
df_long = pd.read_csv('derivatives/preprocessing/dataframe_group.csv')
df_long.head()
| participant | session | age | left-handed | like this course | category | item | ratings | |
|---|---|---|---|---|---|---|---|---|
| 0 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Intouchables | 1.0 |
| 1 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | James Bond | 1.0 |
| 2 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Forrest Gump | 1.0 |
| 3 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Retired Extremely Dangerous | 1.0 |
| 4 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Imitation Game | 1.0 |
As it’s been a few days, we will briefly summarize the data as function of category again:
for index, df in df_long.groupby('category'):
print('Showing information for subdataframe: %s' %index)
print(df['ratings'].describe())
Showing information for subdataframe: animal
count 90.000000
mean 5.701111
std 1.187184
min 2.400000
25% 4.985000
50% 5.940000
75% 6.980000
max 7.000000
Name: ratings, dtype: float64
Showing information for subdataframe: movie
count 255.000000
mean 4.806118
std 1.795347
min 1.000000
25% 4.000000
50% 5.000000
75% 6.260000
max 7.000000
Name: ratings, dtype: float64
Showing information for subdataframe: snack
count 225.000000
mean 5.143822
std 1.633625
min 1.000000
25% 4.000000
50% 5.680000
75% 6.460000
max 7.000000
Name: ratings, dtype: float64
Great! With these basics set, we can continue and start thinking about the potential goal of the analyses.
Recap - What is the goal of the data analyzes¶
There obviously many different routes we could pursue when it comes to analyzing data. Ideally, we would know that before starting (pre-registration much?) but we all know how these things go… For the dataset we aimed at the following, with steps in () indicating operations we already conducted:
(read in single participant data)
(explore single participant data)
(extract needed data from single participant data)
(convert extracted data to more intelligible form)
(repeat for all participant data)
(combine all participant data in one file)
(explore data from all participants)
(general overview)
basic plots
analyze data from all participant
descriptive stats
inferential stats
Nice, that’s a lot. The next step on our list would be data explorations by means of data visualization which will also lead to data analyzes.
Recap - how will the respective steps be implemented¶
After creating some sort of outline/workflow, we though about the respective steps in more detail and set overarching principles. Regarding the former, we also gathered a list of potentially useful python modules to use. Given the pointers above, this entailed the following:
numpy and pandas for data wrangling/exploration
matplolib, seaborn and plotly for data visualization
pingouin and statsmodels for data analyzes/stats
Regarding the second, we went back to standards and principles concerning computational work:
use a dedicated computing environment
provide all steps and analyzes in a reproducible form
nothing will be done manually, everything will be coded
provide as much documentation as possible
Important: these aspects should be followed no matter what you’re working on!
So, after “getting ready” and conducted the first set of processing steps, it’s time to continue via basic data visualization.
Basic data visualization¶
Given that we already explored our data a bit more, including the basic descriptive statistics and data types, we will go one step further and continue this process via basic data visualization to get a different kind of overview that can potentially indicate important aspects concerning data analyses. As mentioned above, we will do so via the following steps, addressing different aspects of data visualization. Throughout each, we will get to know respective python modules and functions.
Underlying principles
“standard” plots
Going further with advanced plots
Underlying principles¶
When talking about visualization one might want to differentiate data exploration and analyses but one can actually drastically influence the other. Here, we are going to check both, that is facilitating data understanding in many ways and creating high quality results figures.
Unsurprisingly, python is nothing but fantastic when it comes to data visualization:
pythonprovides a wide array of optionsLow-level and high-level plotting
APIsstatic
imagesvs.HTMLoutput vs.interactive plotsdomain-general and domain-specific packages
optimal visualization environment as it’s both efficient and flexible
produce off-the-shelf high-quality plots very quickly
with more effort, gives you full control over the plot
While python has a large amount of amazing modules targetting data visualization, we are going to utilize the three most common and general ones, as they provide the basis for everything else going further:
The first two produce static images and the last one HTML outputs and allow much more interactive plots. We will talk about each one as we go along.
matplotlib¶
the most widely-used
pythonplotting libraryinitially modeled on
MATLAB’s plotting systemdesigned to provide complete control over a
plot

matplotlib and all other high-level APIs that build upon it operate on underlying principles and respective parts:
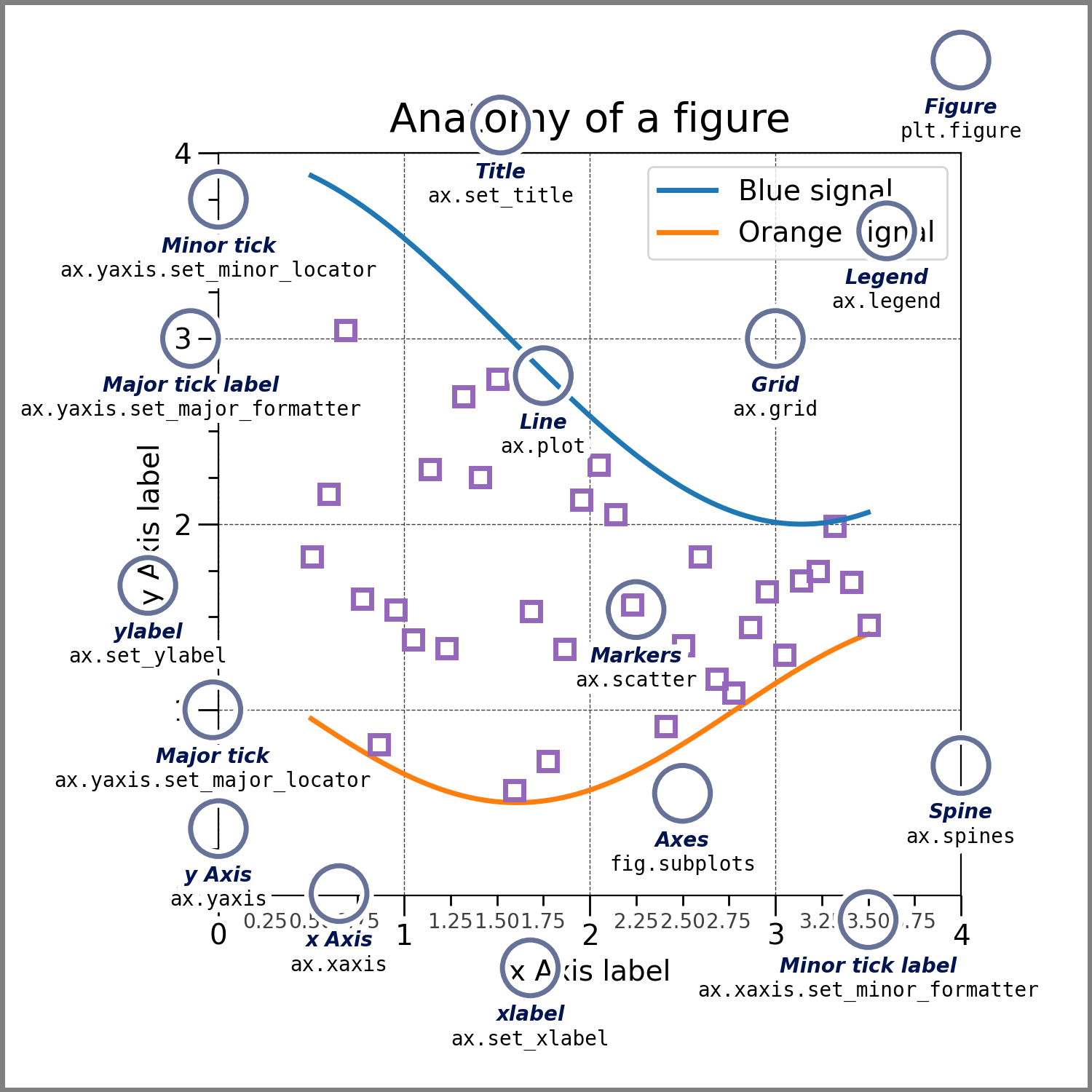
In the most basic sense matplotlib graphs your data on Figures (e.g., windows, Jupyter widgets, etc.), each of which can contain one or more Axes, an area where points can be specified in terms of x-y coordinates (or theta-r in a polar plot, x-y-z in a 3D plot, etc.).
figuresthe entire
graphickeep track of everything therein (
axes,titles,legends, etc.)
axesusually contains two or three
axis objectsincludes
title,x-label,y-label
axisticksandtick labelsto providescalefordata
artisteverything visible on the
figure:text,lines,patches, etc.drawn to the
canvas
A bit too “theoretical”, eh? Let’s dive in and create some plots!
But before we start, two important points to remember: when plotting in jupyter notebooks, make sure to run the %matplotlib inline magic before your first graphic which results in the graphics being embedded in the jupyter notebook and not in the digital void. (NB: this is true for most but not all plotting modules/functions.)
%matplotlib inline
When using matplotlib you can choose between explicitly creating Figures and axes or use the plt interface to automatically create and manage them, as well as adding graphics. Quite often you might want to use the latter.
import matplotlib.pyplot as plt
standard plots¶
Obviously, matplotlib comes with support for all the “standard plots” out there: barplots, scatterplots, histograms, boxplots, errorbars, etc. . For a great overview on what’s possible, make sure to check the gallery of the matplotlib documentation. For now, we are going to start simply…how about some univariate data visualization, e.g. a scatterplot?
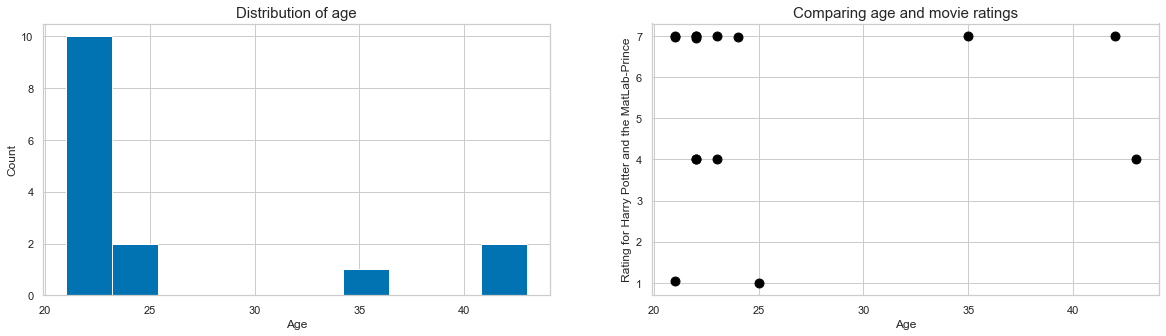
For example, we are interested in the distribution of age in our dataset. Using matplotlib, we need to create a figure and draw something inside. As our data is in long-format we have to initially extract a list containing the age of each participant only once, for example using list comprehension.
plt.figure(figsize=(10, 5))
plt.hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
(array([10., 2., 0., 0., 0., 0., 1., 0., 0., 2.]),
array([21. , 23.2, 25.4, 27.6, 29.8, 32. , 34.2, 36.4, 38.6, 40.8, 43. ]),
<BarContainer object of 10 artists>)
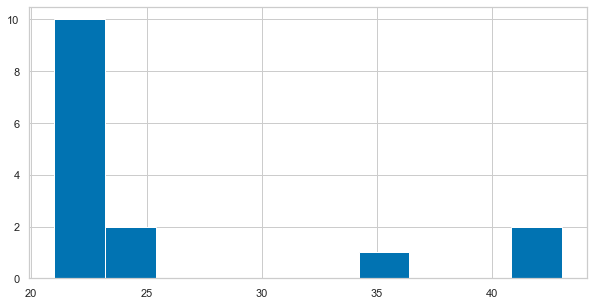
While the information we wanted is there, the plot itself looks kinda cold and misses a few pieces to make it intelligible, e.g. axes labels and a title. This can easily be added via matplotlib’s plt interface.
plt.figure(figsize=(10, 5))
plt.hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
plt.xlabel('Age', fontsize=12)
plt.ylabel('Count', fontsize=12)
plt.title('Distribution of age', fontsize=15);
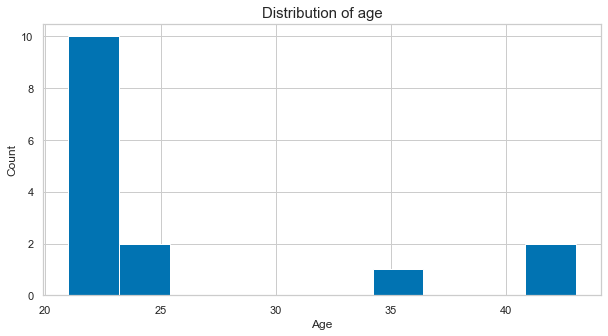
We could also add a grid to make it easier to situate the given values:
plt.figure(figsize=(10, 5))
plt.hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
plt.xlabel('Age', fontsize=12)
plt.ylabel('Count', fontsize=12)
plt.title('Distribution of age', fontsize=15);
plt.grid(True)
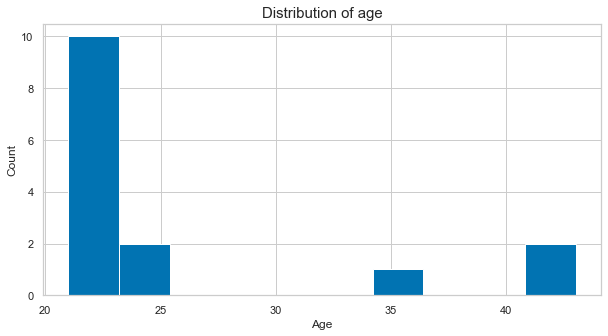
Seeing this distribution of age, we could also have a look how it might interact with certain ratings, i.e. do younger participants rate some of the items different than older participants. Thus, we would create a bivariate visualization with linear data. As an example, let’s look at movies:
df_long[df_long['category']=='movie']['item'].unique()
array(['The Intouchables', 'James Bond', 'Forrest Gump',
'Retired Extremely Dangerous', 'The Imitation Game',
'The Philosophers', 'Call Me by Your Name', 'Shutter Island',
'Love actually', 'The Great Gatsby', 'Interstellar', 'Inception',
'Lord of the Rings - The Two Towers', 'Fight Club', 'Harry Potter',
'Harry Potter and the MatLab-Prince', 'Shindlers List'],
dtype=object)
'Harry Potter and the MatLab-Prince' might be interesting here! Matplotlib’s scatter function comes in handy:
age_list = [df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()]
hp_matlab_ratings = df_long[(df_long['category']=='movie') & (df_long['item']=='Harry Potter and the MatLab-Prince')]['ratings']
plt.figure(figsize=(10, 5))
plt.scatter(age_list, hp_matlab_ratings)
plt.xlabel('Age', fontsize=12)
plt.ylabel('Rating for Harry Potter and the MatLab-Prince', fontsize=12)
plt.title('Comparing age and movie ratings', fontsize=15);
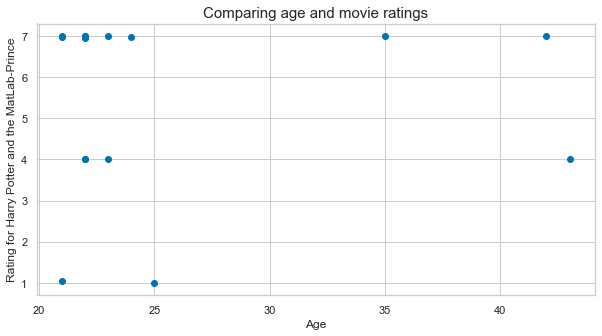
Looks like this movie is somewhat liked by various age groups…To get an idea of the ratings of the other movies let’s extract:
list_ratings = []
for idx, df in df_long[df_long['category']=='movie'].groupby('item'):
list_ratings.append(df['ratings'])
and then plot them via a boxplot()
plt.figure(figsize=(20, 5))
plt.boxplot(list_ratings)
plt.xticks(ticks=range(1,18), labels=df_long[df_long['category']=='movie']['item'].unique())
plt.xticks(rotation = 90)
plt.xlabel('Movies', fontsize=15)
plt.ylabel('Rating', fontsize=15)
plt.title('Rating per movie', fontsize=20);
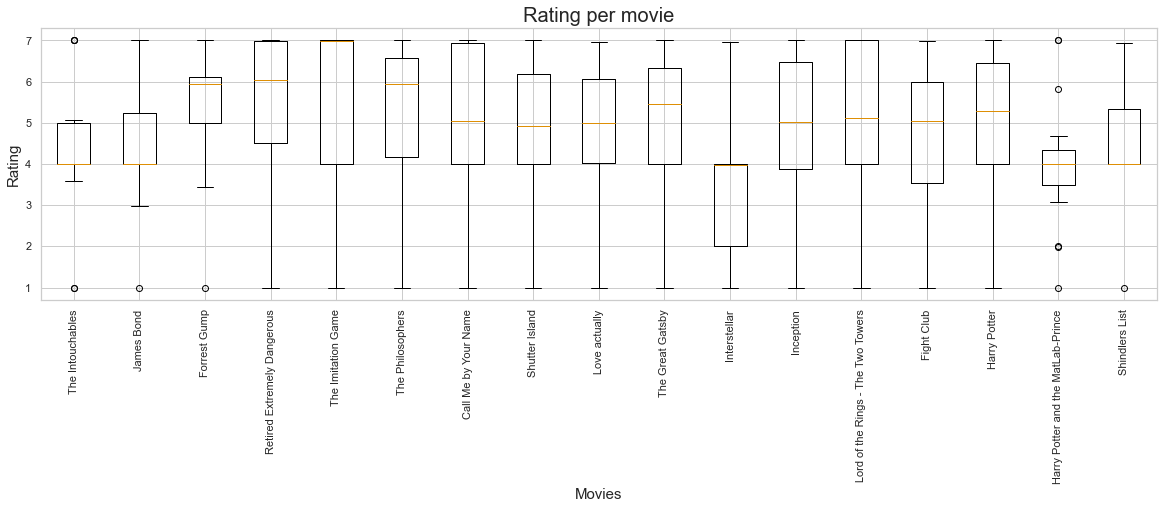
Sometimes, we might want to have different subplots within one main plot. Using matplotlib’s subplots function makes this straightforward via two options: creating a subplot and adding the respective graphics or creating multiple subplots and adding the respective graphics via the axes. Let’s check the first option:
plt.subplot(1, 2, 1)
plt.hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
plt.xlabel('Age', fontsize=12)
plt.ylabel('Count', fontsize=12)
plt.title('Distribution of age', fontsize=15);
plt.grid(True)
plt.subplots_adjust(right=4.85)
plt.subplot(1, 2, 2)
plt.scatter(age_list, hp_matlab_ratings)
plt.xlabel('Age', fontsize=12)
plt.ylabel('Rating for Harry Potter and the MatLab-Prince', fontsize=12)
plt.title('Comparing age and movie ratings', fontsize=15);
plt.show()
Hm, kinda ok but we would need to adapt the size and spacing. This is actually easier using the second option, subplots(), which is also recommended by the matplotlib community:
fig, axs = plt.subplots(1, 2, figsize=(20, 5))
axs[0].hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
axs[0].set_xlabel('Age', fontsize=12)
axs[0].set_ylabel('Count', fontsize=12)
axs[0].set_title('Distribution of age', fontsize=15);
axs[0].grid(True)
axs[1].scatter(age_list, hp_matlab_ratings)
axs[1].set_xlabel('Age', fontsize=12)
axs[1].set_ylabel('Rating for Harry Potter and the MatLab-Prince', fontsize=12)
axs[1].set_title('Comparing age and movie ratings', fontsize=15);
As matplotlib provides access to all parts of a figure, we could furthermore adapt various aspects, e.g. the color and size of the drawn markers.
fig, axs = plt.subplots(1, 2, figsize=(20, 5))
axs[0].hist([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
axs[0].set_xlabel('Age', fontsize=12)
axs[0].set_ylabel('Count', fontsize=12)
axs[0].set_title('Distribution of age', fontsize=15);
axs[0].grid(True)
axs[1].scatter(age_list, hp_matlab_ratings, c='black', s=80)
axs[1].set_xlabel('Age', fontsize=12)
axs[1].set_ylabel('Rating for Harry Potter and the MatLab-Prince', fontsize=12)
axs[1].set_title('Comparing age and movie ratings', fontsize=15);
This provides just a glimpse but matplotlib is infinitely customizable, thus as in most modern plotting environments, you can do virtually anything. The problem is: you just have to be willing to spend enough time on it. Lucky for us and everyone else there are many modules/libraries that provide a high-level interface to matplotlib. However, before we check one of them out we should quickly summarize pros and cons of matplotlib.
Pros¶
provides low-level control over virtually every element of a
plotcompletely
object-oriented API;plotcomponents can be easily modifiedclose integration with
numpyextremely active community
tons of functionality (
figure compositing,layering,annotation,coordinate transformations,color mapping, etc.)
Cons¶
steep learning curve
APIis extremely unpredictable–redundancy and inconsistency are commonsome simple things are hard; some complex things are easy
lacks systematicity/organizing
syntax–everyplotis its own little worldsimple
plotsoften require a lot ofcodedefault
stylesare not optimal
High-level interfaces to matplotlib¶
matplotlibis very powerful and very robust, but theAPIis hit-and-missmany high-level interfaces to
matplotlibhave been writtenabstract away many of the annoying details
best of both worlds: easy generation of
plots, but retainmatplotlib’s powerSeaborn
ggplot
pandas
etc.
many domain-specific
visualizationtools are built onmatplotlib(e.g.,nilearnandmneinneuroimaging)
Going further with advanced plots¶
This also marks the transition to more “advanced plots” as the respective libraries allow you to create fantastic and complex plots with ease!
Seaborn¶
Seaborn abstracts away many of the complexities to deal with such minutiae and provides a high-level API for creating aesthetic plots.
arguably the premier
matplotlibinterfaceforhigh-level plotsgenerates beautiful
plotsin very littlecodebeautiful
stylesandcolor paletteswide range of supported plots
modest support for structured
plotting(viagrids)exceptional documentation
generally, the best place to start when
exploringandvisualizing data(can be quite slow (e.g., with
permutation))
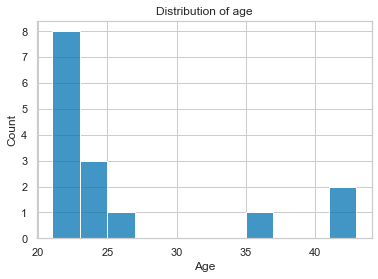
For example, recreating the plots from above is as easy as:
import seaborn as sns
sns.histplot([df_long[df_long['participant']==part]['age'].to_numpy()[0] for part in df_long['participant'].unique()])
plt.xlabel('Age')
plt.title('Distribution of age')
Text(0.5, 1.0, 'Distribution of age')
sns.scatterplot(x=age_list, y=hp_matlab_ratings)
plt.xlabel('Age')
plt.title('Comparing age and movie ratings')
Text(0.5, 1.0, 'Comparing age and movie ratings')
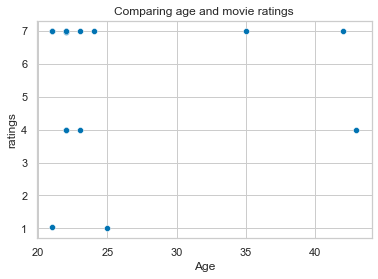
You might wonder: “well, that doesn’t look so different from the plots we created before and it’s also not way faster/easier”.
True that, but so far this based on our data and the things we wanted to plot. Seaborn actually integrates fantastically with pandas dataframes and allows to achieve amazing things rather easily.
Let’s go through some examples!
How about evaluating ratings as function of age, separated by category (movies, snacks, animals)? Sounds wild? Using seaborn’s pairplot this achieved with just one line of code:
sns.pairplot(df_long[['age', 'category', 'item', 'ratings']], hue='category')
<seaborn.axisgrid.PairGrid at 0x167550d90>
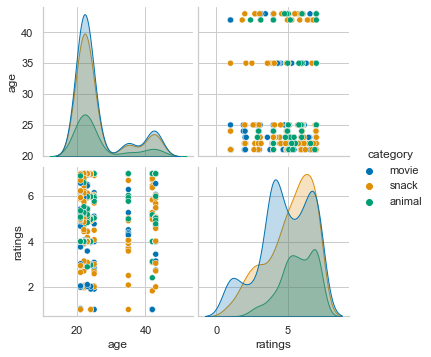
Or how about rating of animals as a function of age, separately for each animal? Same approach, but restricted to a subdataframe that only contains the ratings of the animal category!
sns.pairplot(df_long[df_long['category']=='animal'][['age', 'item', 'ratings']], hue='item')
<seaborn.axisgrid.PairGrid at 0x16774cd30>
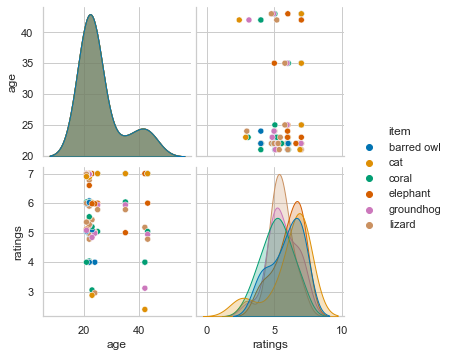
Assuming we want to check ratings of the movie category further, we will create a respective subdataframe.
df_movies = df_long[df_long['category']=='movie']
df_movies.head(n=10)
| participant | session | age | left-handed | like this course | category | item | ratings | |
|---|---|---|---|---|---|---|---|---|
| 0 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Intouchables | 1.0 |
| 1 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | James Bond | 1.0 |
| 2 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Forrest Gump | 1.0 |
| 3 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Retired Extremely Dangerous | 1.0 |
| 4 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Imitation Game | 1.0 |
| 5 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Philosophers | 1.0 |
| 6 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Call Me by Your Name | 1.0 |
| 7 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Shutter Island | 1.0 |
| 8 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | Love actually | 1.0 |
| 9 | Smitty_Werben_Jagger_Man_Jensen#1 | 1.0 | 42.0 | False | No | movie | The Great Gatsby | 1.0 |
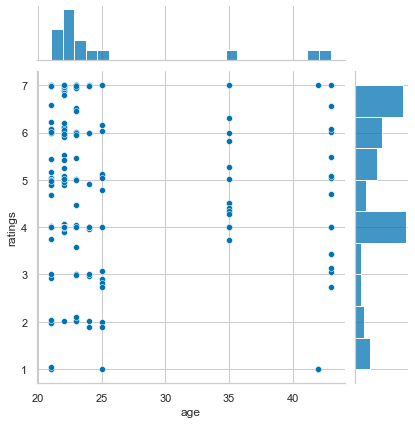
And can now make use of the fantastic pandas - seaborn friendship. For example, let’s go back to the scatterplot of age and ratings we did before. How could we improve this plot? Maybe adding the distribution of each variable to it? That’s easily done via jointplot():
sns.jointplot(x='age', y='ratings', data=df_movies)
<seaborn.axisgrid.JointGrid at 0x1679937c0>
Wouldn’t it be cool if we could also briefly explore if there might be some statistically relevant effects going on here? Say no more, as we can add a regression to the plot via setting the kind argument to reg:
sns.jointplot(x='age', y='ratings', data=df_movies, kind='reg')
<seaborn.axisgrid.JointGrid at 0x167a92130>
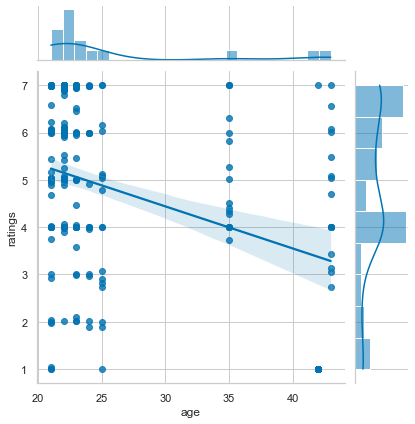
Chances are we see another manifestation of early onset grumpiness here and thus might want to spend a closer look on the ratings for each movie. One possibility to do so, could be a boxplot():
plt.figure(figsize=(10,10))
sns.boxplot(x='ratings', y='item', data=df_movies, palette="vlag")
<AxesSubplot:xlabel='ratings', ylabel='item'>
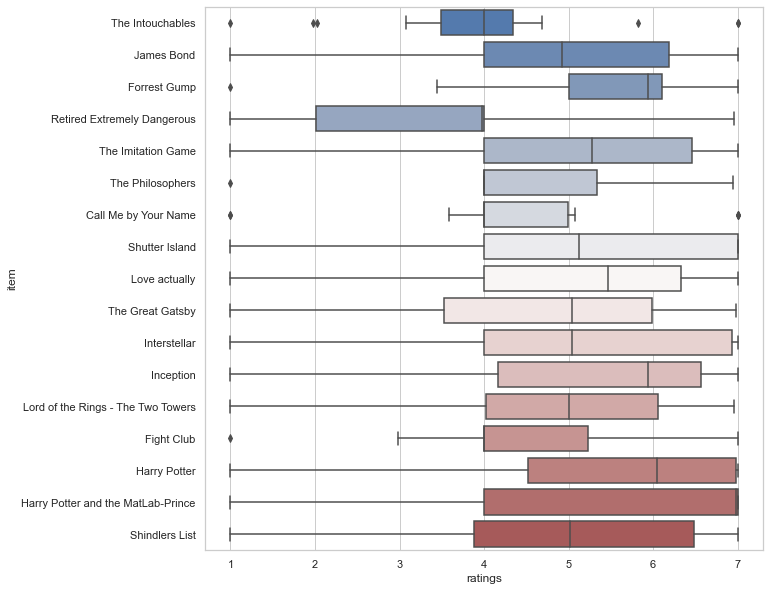
However, we know that boxplots have their fair share of problems…given that they show summary statistics, clusters and multimodalities are hidden..

That’s actually one important aspect everyone should remember concerning data visualization, no matter if for exploration or analyses: show as much data and information as possible! With seaborn we can easily address this via adding individual data points to our plot via stripplot():
plt.figure(figsize=(10,10))
sns.boxplot(x='ratings', y='item', data=df_movies, palette="vlag")
sns.stripplot(x='ratings', y='item', data=df_movies, color='black')
<AxesSubplot:xlabel='ratings', ylabel='item'>
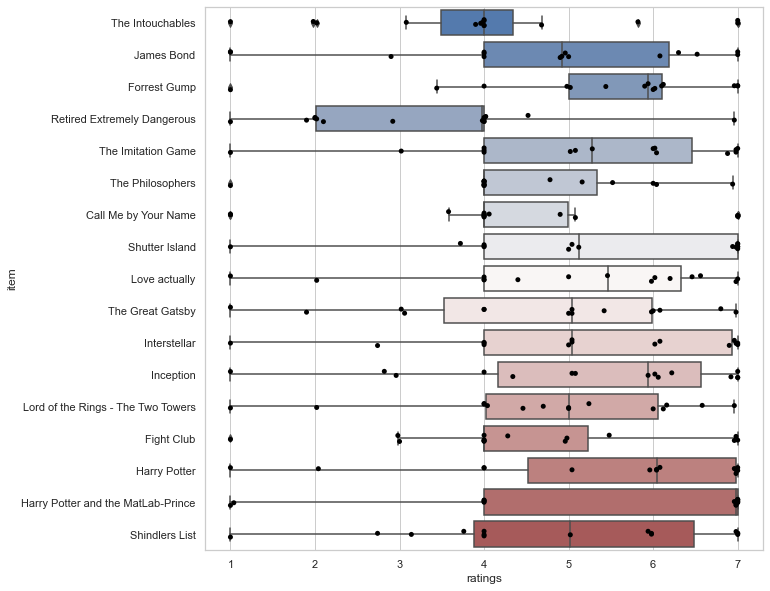
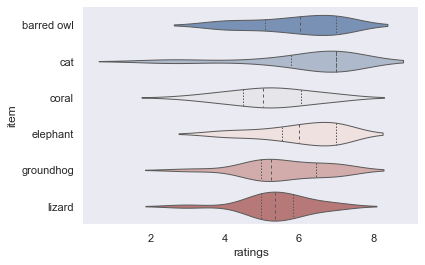
Ah yes, that’s better! Seeing the individual data points, we might want to check the respective distributions. Using seaborn’s violinplot(), this is done in no time.
plt.figure(figsize=(10,10))
sns.violinplot(data=df_movies, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
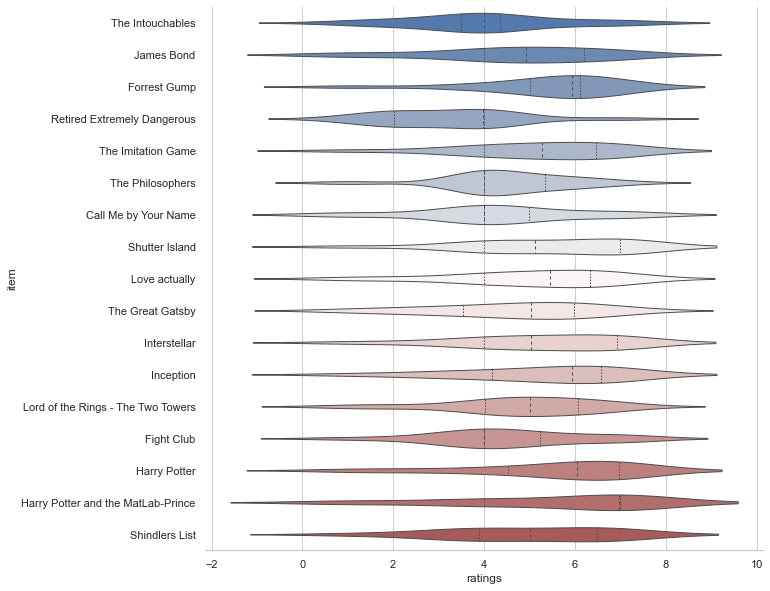
As you might have seen, we also adapted the style of our plot a bit via sns.despine() which removed the y axis spine. This actually outlines another important point: seaborn is fantastic when it comes to customizing plots with little effort (i.e. getting rid of many lines of matplotlib code). This includes “themes, contexts, colormaps among other things. The subdataframe including animal ratings might be a good candidate to explore these aspects.
df_animals = df_long[df_long['category']=='animal']
Starting with themes, which set a variety of aesthetic related factors, including background color, grids, etc., here are some very different examples showcasing the whitegrid style
sns.set_style("whitegrid")
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
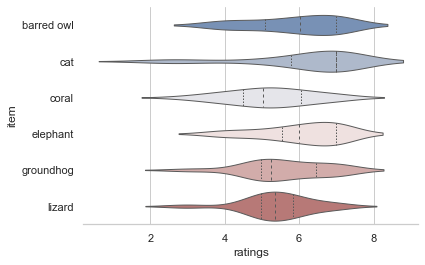
and the dark style:
sns.set_style("dark")
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
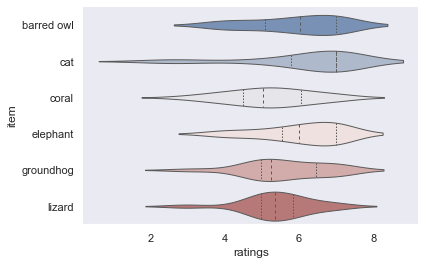
While this is already super cool, seaborn goes one step further and even let’s you define the context for which your figure is intended and adapts it accordingly. For example, there’s a big difference if you want to include your figure in a poster:
sns.set_style('whitegrid')
sns.set_context("poster")
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
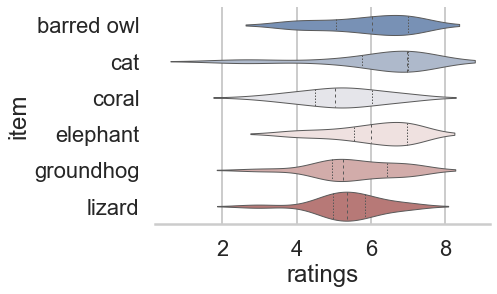
or a talk:
sns.set_context("talk")
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
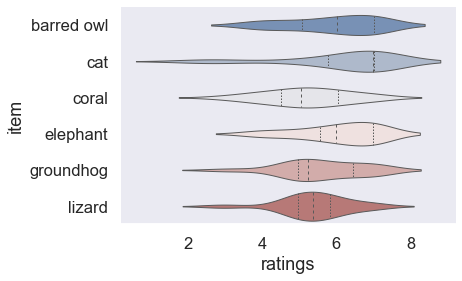
or a paper:
sns.set_context("paper")
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
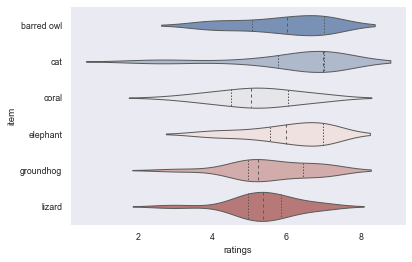
No matter the figure and context, another very crucial aspect everyone should always look out for is the colormap or colorpalette! Some of the most common ones are actually suboptimal in multiple regards. This entails a misrepresentation of data:
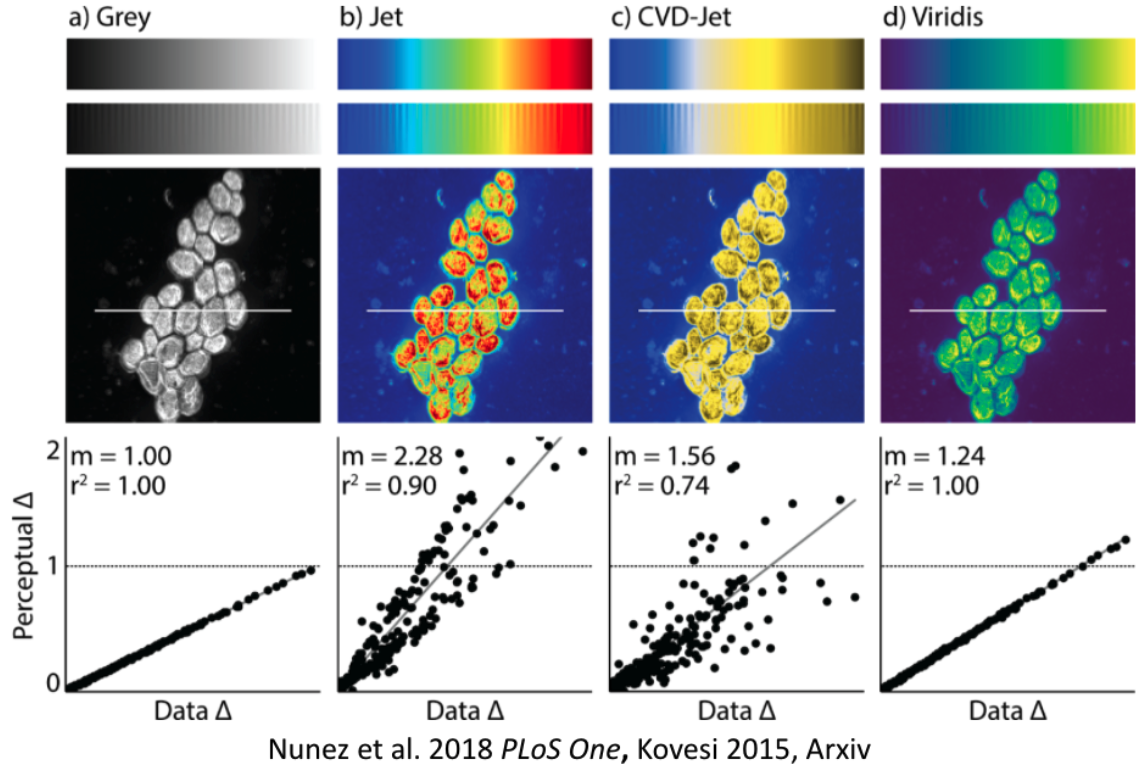
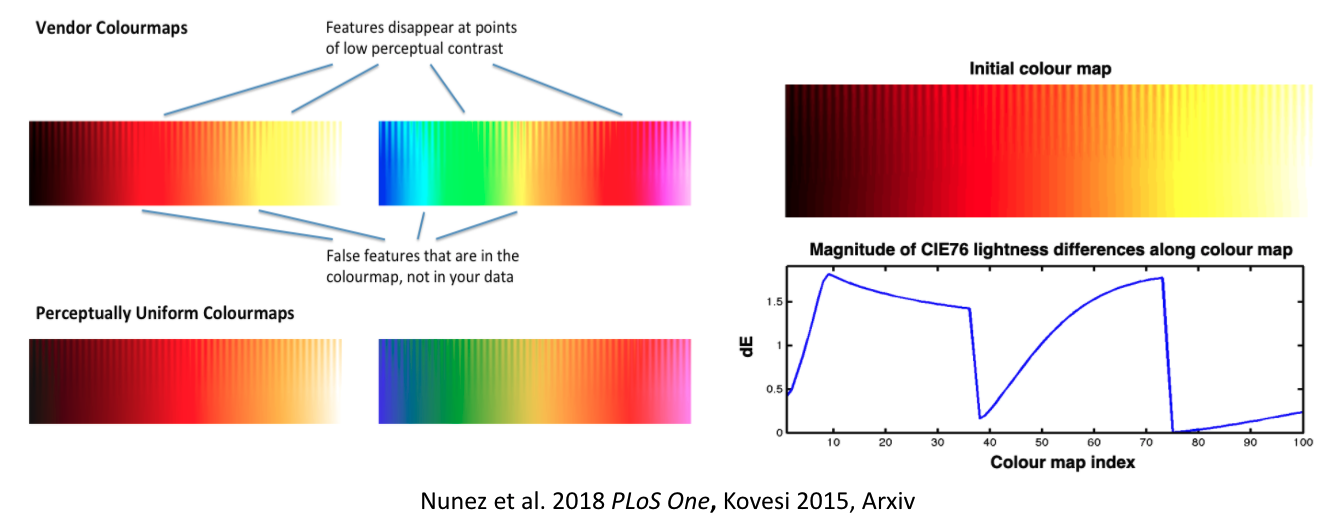
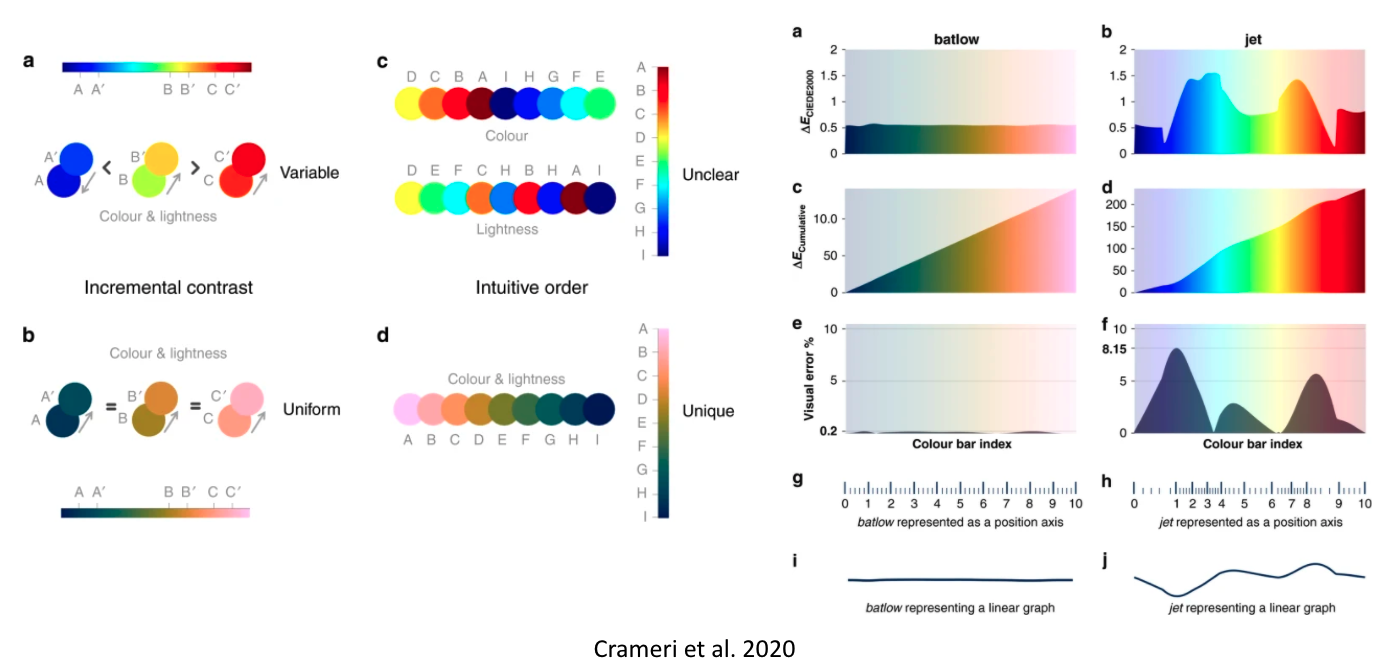
It gets even worse: they don’t work for people with color vision deficiencies!
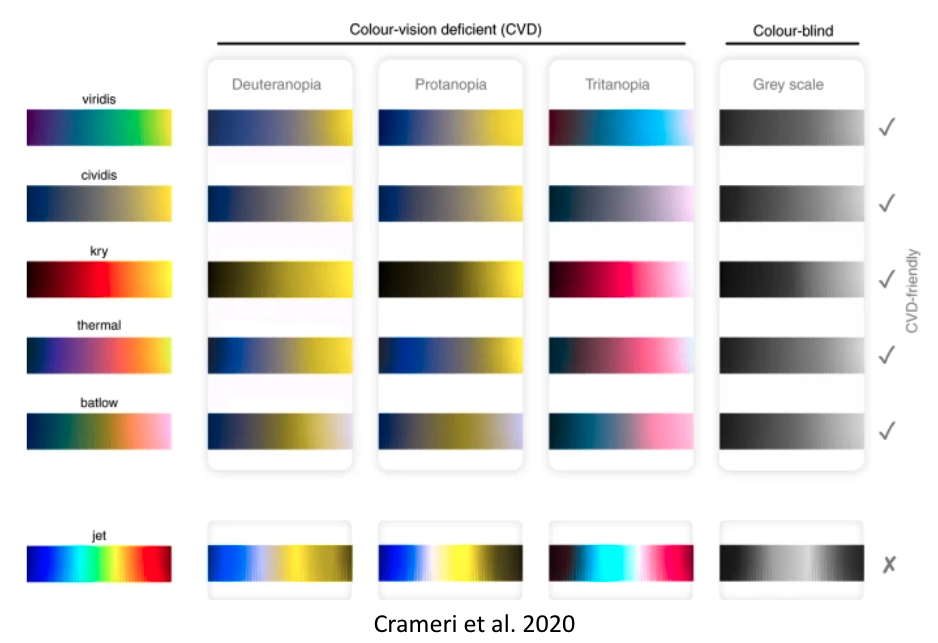
That’s obviously not ok and we/you need to address this! With seaborn, some of these important aspects are easily addressed. For example, via setting the colorpalette to colorblind:
sns.set_context("notebook")
sns.set_palette('colorblind')
sns.violinplot(data=df_animals, x="ratings", y="item", inner="quart", linewidth=1, palette='vlag')
sns.despine(left=True)
or using one of the suitable color palettes that also address the data representation problem, i.e. perceptually uniform color palettes:
sns.color_palette("flare", as_cmap=True)
sns.color_palette("crest", as_cmap=True)
sns.cubehelix_palette(as_cmap=True)
sns.cubehelix_palette(start=.5, rot=-.5, as_cmap=True)
Let’s see a few of those in action, for example within a heatmap that displays the correlation between ratings of movies. For this, we need to reshape our data back to wide-format which is straightforward using pandas’ pivot function.
df_movies_wide = pd.pivot(df_movies[['participant', 'category', 'item', 'ratings']], index='participant', columns=['category', 'item'])
df_movies_wide.columns = df_movies['item'].unique()
Then we can use another built-in function of pandas dataframes: .corr(), which computes a correlation between all columns:
plt.figure(figsize=(10,7))
sns.heatmap(df_movies_wide.corr(), xticklabels=False, cmap='rocket')
<AxesSubplot:>
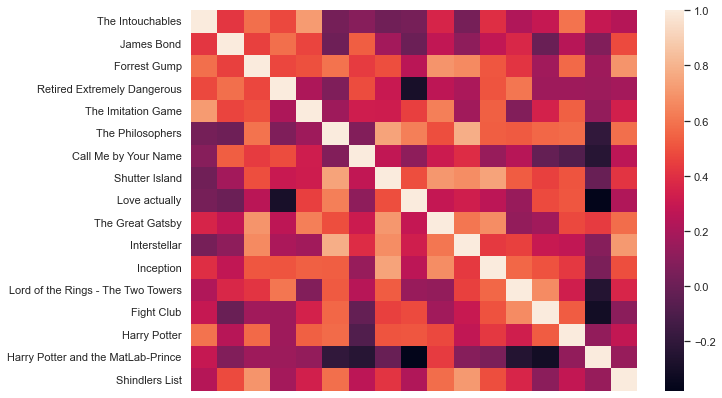
Nice, how does the crest palette look?
plt.figure(figsize=(10,7))
sns.heatmap(df_movies_wide.corr(), xticklabels=False, cmap='crest')
<AxesSubplot:>
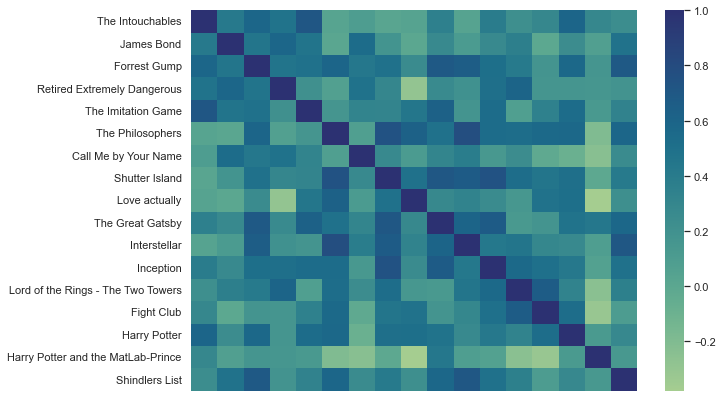
Also fantastic! However, it’s easy to get fooled by beautiful graphics and given that our values include negative and positive numbers, we should use a diverging color palette. While we’re at it, we also change heatmap to clustermap!
plt.figure(figsize=(10,7))
sns.clustermap(df_movies_wide.corr(), xticklabels=False, cmap='vlag', center=0)
<seaborn.matrix.ClusterGrid at 0x168762d00>
<Figure size 720x504 with 0 Axes>
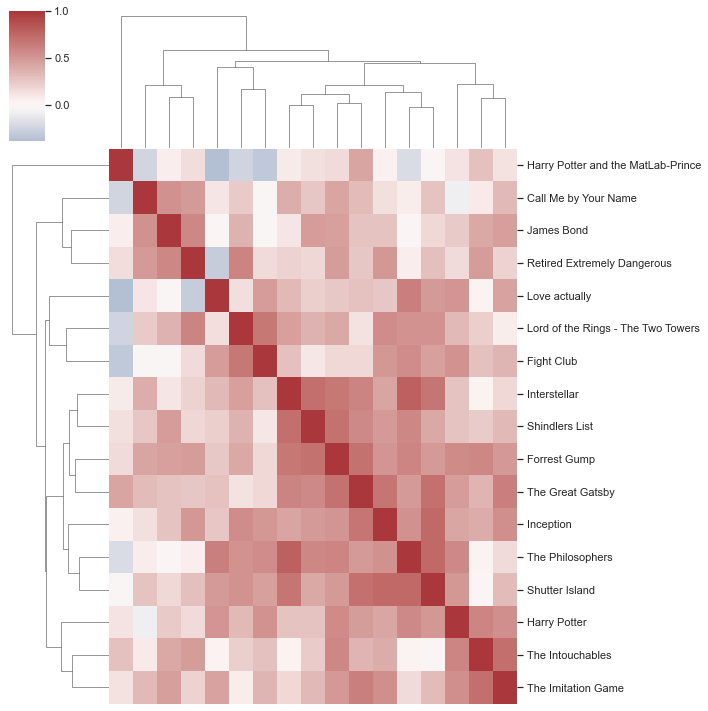

However, to be on the safe side, please also check your graphics for the mentioned points, e.g. via tools like Color Oracle that let you simulate color vision deficiencies!
Make use of amazing resources like the python graph gallery, the data to viz project and the colormap decision tree in Crameri et al. 2020, that in combination allow you to find and use the best graphic and colormap for your data!
And NEVER USE JET!
While the things we briefly explored were already super cool and a lot, we cannot conclude the data visualization section without at least mentioning the up and coming next-level graphics: raincloudplots as they combine various aspects of the things we’ve talked about! In python they are available via the ptitprince library.
# run this cell if you didn't install ptitprince yet
pip install ptitprince
from ptitprince import PtitPrince as pt
f, ax = plt.subplots(figsize=(12, 11))
pt.RainCloud(data = df_long, x = "category", y = "ratings", ax = ax, orient='h')
<AxesSubplot:xlabel='ratings', ylabel='category'>
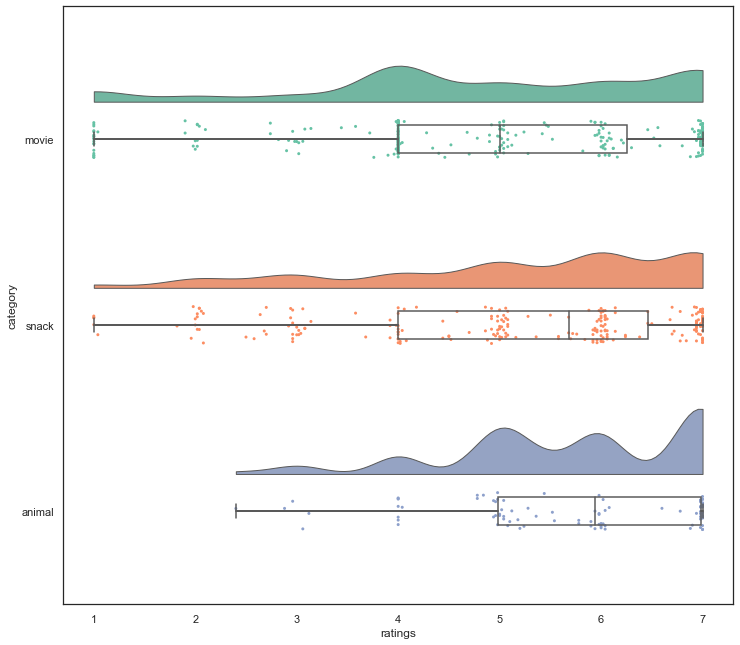
Via our adventures in basic data visualization we actually learned quite a bit about our data and enhanced our understanding obtained through the initial exploration via data handling and wrangling.
There appear to be some interesting effects we should investigate further which leads us to the grand finale: statistical analyses using python!
Statistics in python¶
We’ve reached the final step of our analyses workflow: statistics, i.e. putting things to test via evaluating hypotheses and/or assumptions! As with the previous aspects, python has a lot of libraries and modules for this purpose, some general-domain and some rather tailored to specific data and analyzes (e.g. fMRI, EEG, drift diffusion models, etc.).
Here, we will stick with the general-domain ones as they are already support a huge amount of approaches and are commonly used by the community: pingouin and statsmodels. As before, we will go through several steps and explore respective python modules and functions.
Statistics in python
Descriptive analyses
Inferential analyses
Descriptive analyses¶
Regarding the first aspect, descriptive analyses, we can safely state that we already know how to do this. For example using pandas or numpy and their respective functions:
print(df_animals['ratings'].mean(), df_animals['ratings'].std())
5.7011111111111115 1.187183849761099
import numpy as np
print(np.mean(df_animals['ratings']), np.std(df_animals['ratings']))
5.7011111111111115 1.180569960696428
for idx, df in df_long[['age', 'category', 'ratings']].groupby('category'):
print('Summary statistics for: %s' %idx)
print(df.describe())
Summary statistics for: animal
age ratings
count 90.000000 90.000000
mean 25.866667 5.701111
std 7.360997 1.187184
min 21.000000 2.400000
25% 22.000000 4.985000
50% 22.000000 5.940000
75% 25.000000 6.980000
max 43.000000 7.000000
Summary statistics for: movie
age ratings
count 255.000000 255.000000
mean 25.866667 4.806118
std 7.334383 1.795347
min 21.000000 1.000000
25% 22.000000 4.000000
50% 22.000000 5.000000
75% 25.000000 6.260000
max 43.000000 7.000000
Summary statistics for: snack
age ratings
count 225.000000 225.000000
mean 25.866667 5.143822
std 7.336309 1.633625
min 21.000000 1.000000
25% 22.000000 4.000000
50% 22.000000 5.680000
75% 25.000000 6.460000
max 43.000000 7.000000
Thus, we can skip these aspects here and directly move on! Regarding inferential analyses, we will start with and focus on pingouin!
Unfortunately not those…

but make sure to check https://www.penguinsinternational.org/ to see what you can do to help our amazing friends: the Penguins!
Inferential analyses¶
Of course, exploring and visualizing data is informative and can hint at certain effects but we nevertheless have to run tests and obtain respective statistics to evaluate if there’s “something meaningful”. This is where inferential analyses come into play!
Pingouin¶

Even though comparably new, Pingouin quickly became a staple regarding statistical analyses as it integrates amazingly well with the existing python data science stack, e.g. pandas and numpy. As mentioned in the docs: “Pingouin is designed for users who want simple yet exhaustive stats functions.”, which summarizes it perfectly as it supports the majority of commonly used statistical functions and additionally provides adjacent functionality, e.g. related to test diagnostics and evaluation:
ANOVAs:N-ways,repeated measures,mixed,ancovapairwise post-hocs tests(parametricandnon-parametric) andpairwise correlationsrobust,partial,distanceandrepeated measures correlationslinear/logisticregressionandmediation analysisBayes Factorsmultivariate testsreliabilityandconsistencyeffect sizesandpower analysisparametric/bootstrappedconfidence intervalsaround aneffect sizeor acorrelation coefficientcircular statisticschi-squared testsplotting:Bland-Altman plot,Q-Q plot,paired plot,robust correlation…
When in doubt, make sure to check if pingouin has your back!
One of the first things we explored via the previous analyses steps was if the ratings vary as a function of age. While the plots suggested at some potential effects, we can simply compute their correlation using pingouin’s corr function:
import pingouin as pg
pg.corr(age_list, hp_matlab_ratings)
| n | r | CI95% | p-val | BF10 | power | |
|---|---|---|---|---|---|---|
| pearson | 15 | 0.06325 | [-0.46, 0.56] | 0.822805 | 0.326 | 0.054932 |
Not only do we get the correlation value and p value but also the number of observations, the 95% CI, Bayes Factor and the power. Pretty convenient, eh?
If we now would like to compute the robust biweight midcorrelation, we only need to set the method argument accordingly:
pg.corr(age_list, hp_matlab_ratings, method='bicor')
| n | r | CI95% | p-val | power | |
|---|---|---|---|---|---|
| bicor | 15 | 0.0 | [-0.51, 0.51] | 1.0 | 0.049065 |
Using heatmap and clustermaps we also evaluate the correlation pattern of movie ratings. In order to put some more numbers on that, we can make use of the mentioned pingouin - pandas friendship and compute multiple pairwise correlations based on the columns of a dataframe:
movie_cor = pg.pairwise_corr(df_movies_wide, method='pearson')
movie_cor.head()
| X | Y | method | alternative | n | r | CI95% | p-unc | BF10 | power | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | The Intouchables | James Bond | pearson | two-sided | 15 | 0.419801 | [-0.12, 0.77] | 0.119273 | 0.97 | 0.357333 |
| 1 | The Intouchables | Forrest Gump | pearson | two-sided | 15 | 0.581488 | [0.1, 0.84] | 0.022984 | 3.391 | 0.657831 |
| 2 | The Intouchables | Retired Extremely Dangerous | pearson | two-sided | 15 | 0.474495 | [-0.05, 0.79] | 0.073921 | 1.378 | 0.451380 |
| 3 | The Intouchables | The Imitation Game | pearson | two-sided | 15 | 0.716234 | [0.32, 0.9] | 0.002668 | 19.388 | 0.892110 |
| 4 | The Intouchables | The Philosophers | pearson | two-sided | 15 | 0.032963 | [-0.49, 0.54] | 0.907161 | 0.32 | 0.050651 |
We get a dataframe in return, which we can then use for further analyses. For example, as we did run quite a few tests, we might want to correct our p values for multiple comparisons before we make any claims regarding the rating of certain movies being correlated. Obviously, pingouin also makes this super easy via it’s multicomp function which returns an list with True/False booleans and corrected p values:
movie_cor['p_corr'] = pg.multicomp(movie_cor['p-unc'].to_numpy(), method='fdr_bh')[1]
movie_cor.head()
| X | Y | method | alternative | n | r | CI95% | p-unc | BF10 | power | p_corr | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | The Intouchables | James Bond | pearson | two-sided | 15 | 0.419801 | [-0.12, 0.77] | 0.119273 | 0.97 | 0.357333 | 0.272475 |
| 1 | The Intouchables | Forrest Gump | pearson | two-sided | 15 | 0.581488 | [0.1, 0.84] | 0.022984 | 3.391 | 0.657831 | 0.148849 |
| 2 | The Intouchables | Retired Extremely Dangerous | pearson | two-sided | 15 | 0.474495 | [-0.05, 0.79] | 0.073921 | 1.378 | 0.451380 | 0.209986 |
| 3 | The Intouchables | The Imitation Game | pearson | two-sided | 15 | 0.716234 | [0.32, 0.9] | 0.002668 | 19.388 | 0.892110 | 0.070132 |
| 4 | The Intouchables | The Philosophers | pearson | two-sided | 15 | 0.032963 | [-0.49, 0.54] | 0.907161 | 0.32 | 0.050651 | 0.949030 |
We could now go “classic” and check if any if the corrected p values is below a certain threshold…yes, we can check 0.05 if you want:
movie_cor[movie_cor['p_corr'] <= 0.05]
| X | Y | method | alternative | n | r | CI95% | p-unc | BF10 | power | p_corr |
|---|
Hm, doesn’t like that’s the case. Because we didn’t preregister anything we could start our wild goose chase here. (Obviously you know, that’s neither cool nor the ideal workflow and should be avoided at any cost.)
We could reformulate our analyses to an regression problem and thus run a multiple regression via pingouin’s linear_regression function:
movie_reg = pg.linear_regression(df_movies_wide[['James Bond', 'Forrest Gump']], age_list)
movie_reg
| names | coef | se | T | pval | r2 | adj_r2 | CI[2.5%] | CI[97.5%] | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Intercept | 12.943912 | 6.893094 | 1.877809 | 0.084918 | 0.263854 | 0.141163 | -2.074848 | 27.962673 |
| 1 | James Bond | 1.382941 | 1.106310 | 1.250048 | 0.235111 | 0.263854 | 0.141163 | -1.027501 | 3.793383 |
| 2 | Forrest Gump | 1.206331 | 1.324423 | 0.910835 | 0.380315 | 0.263854 | 0.141163 | -1.679338 | 4.092000 |
Which would again provide a very exhaustive and intelligible output that can easily be utilized further.
One of the outputs of the regression reminds us of “a very low hanging fruit” we haven’t checked yet: T-Tests! For example, we could check if the ratings between two movies are different. However, before we can do this, isn’t there something we should do?
That’s right: testing statistical premises/test assumptions! In other words: we need to evaluate if the assumptions/requirements for a parametric test, like the T-Test are fulfilled or if we need to apply a non-parametric test.
At first, we are going to test the distribution of our data. Often folks assume that their data follows a gaussian distribution, which allows for parametric tests to be run. Nevertheless it is essential to first test the distribution of your data to decide if the assumption of normally distributed data holds, if this is not the case we would have to switch to non-parametric tests.
With pingouin this is again only one brief function away. Here specifically, the normality() function that implements the Shapiro Wilk normality test:
pg.normality(df_movies_wide['Interstellar'])
| W | pval | normal | |
|---|---|---|---|
| Interstellar | 0.886377 | 0.059126 | True |
pg.normality(df_movies_wide['Inception'])
| W | pval | normal | |
|---|---|---|---|
| Inception | 0.890249 | 0.067643 | True |
“So far so good”. Next, let’s have a look at homoscedasticity, i.e. checking the variance of ratings of the two movies in question. (NB: depending on the function and data at hand you can either directly operate on dataframes or need to provide lists of arrays. Always make sure to check the documentation!)
pg.homoscedasticity([df_movies_wide['Interstellar'].to_numpy(),df_movies_wide['Inception'].to_numpy()])
| W | pval | equal_var | |
|---|---|---|---|
| levene | 0.000727 | 0.978687 | True |
Seems like we could go ahead and compute a T-Test.
By now you might know that pingouin obviously has a dedicated function for this and of course you’re right: ttest:
pg.ttest(df_movies_wide['Interstellar'], df_movies_wide['Inception'], paired=True)
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 0.048337 | 14 | two-sided | 0.962131 | [-1.04, 1.09] | 0.013285 | 0.263 | 0.050264 |
Comparable to the correlation analyses, we get quite a few informative and helpful outputs beyond “classic test statistics”!
If we however wanted to run a non-parametric test, here a wilcoxon test, we can do so without problems:
pg.wilcoxon(df_movies_wide['Interstellar'], df_movies_wide['Inception'])
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/scipy/stats/morestats.py:3141: UserWarning: Exact p-value calculation does not work if there are ties. Switching to normal approximation.
warnings.warn("Exact p-value calculation does not work if there are "
| W-val | alternative | p-val | RBC | CLES | |
|---|---|---|---|---|---|
| Wilcoxon | 41.5 | two-sided | 0.806708 | -0.087912 | 0.482222 |
While we could now go through each comparison of interest, we might want to approach the analyses from a different angle. For example, starting with an anova comparing ratings between categories:
pg.rm_anova(data=df_long, dv='ratings', within='category', subject='participant', detailed=True)
| Source | SS | DF | MS | F | p-unc | np2 | eps | |
|---|---|---|---|---|---|---|---|---|
| 0 | category | 6.128143 | 2 | 3.064071 | 5.685984 | 0.008464 | 0.288834 | 0.865891 |
| 1 | Error | 15.088683 | 28 | 0.538882 | NaN | NaN | NaN | NaN |
Usually the next step would be to run post-hoc tests contrasting each pair of categories. Once more pingouin comes to the rescue and implements this in just one function within which we can additionally define if we want to run a parametric test, if p values should be adjusted and what kind of effect size we would like to have:
pg.pairwise_ttests(data=df_long, dv='ratings', within='category', subject='participant',
parametric=True, padjust='fdr_bh', effsize='cohen')
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | p-corr | p-adjust | BF10 | cohen | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | category | animal | movie | True | True | 3.800629 | 14.0 | two-sided | 0.001948 | 0.005845 | fdr_bh | 21.898 | 0.984213 |
| 1 | category | animal | snack | True | True | 2.273248 | 14.0 | two-sided | 0.039291 | 0.058936 | fdr_bh | 1.854 | 0.835592 |
| 2 | category | movie | snack | True | True | -1.067915 | 14.0 | two-sided | 0.303628 | 0.303628 | fdr_bh | 0.427 | -0.397164 |
The same can also be done within a given category, comparing the corresponding items:
pg.pairwise_ttests(data=df_animals, dv='ratings', within='item', subject='participant',
parametric=True, padjust='fdr_bh', effsize='cohen')
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | p-corr | p-adjust | BF10 | cohen | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | item | barred owl | cat | True | True | -0.821873 | 14.0 | two-sided | 0.424926 | 0.573550 | fdr_bh | 0.352 | -0.166131 |
| 1 | item | barred owl | coral | True | True | 1.933279 | 14.0 | two-sided | 0.073684 | 0.224741 | fdr_bh | 1.144 | 0.593782 |
| 2 | item | barred owl | elephant | True | True | -0.340680 | 14.0 | two-sided | 0.738403 | 0.791146 | fdr_bh | 0.276 | -0.144527 |
| 3 | item | barred owl | groundhog | True | True | 1.060789 | 14.0 | two-sided | 0.306744 | 0.460116 | fdr_bh | 0.424 | 0.255968 |
| 4 | item | barred owl | lizard | True | True | 1.755625 | 14.0 | two-sided | 0.100998 | 0.224741 | fdr_bh | 0.905 | 0.451942 |
| 5 | item | cat | coral | True | True | 2.197715 | 14.0 | two-sided | 0.045294 | 0.224741 | fdr_bh | 1.659 | 0.676153 |
| 6 | item | cat | elephant | True | True | 0.121288 | 14.0 | two-sided | 0.905187 | 0.905187 | fdr_bh | 0.264 | 0.049262 |
| 7 | item | cat | groundhog | True | True | 1.852810 | 14.0 | two-sided | 0.085105 | 0.224741 | fdr_bh | 1.027 | 0.388575 |
| 8 | item | cat | lizard | True | True | 1.781298 | 14.0 | two-sided | 0.096562 | 0.224741 | fdr_bh | 0.935 | 0.556165 |
| 9 | item | coral | elephant | True | True | -2.240984 | 14.0 | two-sided | 0.041757 | 0.224741 | fdr_bh | 1.768 | -0.772576 |
| 10 | item | coral | groundhog | True | True | -1.262752 | 14.0 | two-sided | 0.227306 | 0.426199 | fdr_bh | 0.513 | -0.354908 |
| 11 | item | coral | lizard | True | True | -0.626458 | 14.0 | two-sided | 0.541095 | 0.624341 | fdr_bh | 0.312 | -0.193253 |
| 12 | item | elephant | groundhog | True | True | 1.146601 | 14.0 | two-sided | 0.270766 | 0.451277 | fdr_bh | 0.458 | 0.420019 |
| 13 | item | elephant | lizard | True | True | 1.733964 | 14.0 | two-sided | 0.104879 | 0.224741 | fdr_bh | 0.88 | 0.640226 |
| 14 | item | groundhog | lizard | True | True | 0.761768 | 14.0 | two-sided | 0.458840 | 0.573550 | fdr_bh | 0.338 | 0.188385 |
Isn’t pingouin just fantastic? Especially considering that the many things we explored only cover a small percentage of available functionality as we didn’t even look at e.g. mediation analyses, contingency analyses, multivariate tests, reliability, etc. . However, you should have got an idea how powerful and versatile pingouin is for the majority of analyses you commonly run.
NB 1: A small note before we continue: a lot of these “standard analyses” and more are also implemented in the scipy.stats module which is might be interesting if there’s something you want to run that is implemented pingouin.
NB 2: we focused on pingouin because of it’s large amount of functions, great integration with pandas and high-level API.
Even though it’s a virtual class I can literally hear folks yell: “What about formulas? I need to define formulas!”.
Don’t you worry a thing: while this is (yet) not possible in pingouin, there’s of course a different python module that can do these things. Open your hearts for statsmodels!
Statsmodels¶
statsmodels (sorry no animal pun, don’t know why) is yet another great python module for statistical analyses. In contrast to pingouin it focuses on rather complex and more specialized analyses:
regressiondiscrete choice modelsGeneralized Estimating Equationsmeta-analysestime-series,forecastingstate space modelsetc.
In order to provide a brief glimpse of how statsmodels can be used to define and apply statistical models via formulas, we will check two examples and while doing so ignoring the reasonableness. At first we will import the formula.api:
import statsmodels.formula.api as smf
Which can then be used to define and run statistical models. Independent of the specific model the outline is roughly identical: a model is defined via a string e.g. referencing the columns/variables of a dataframe and then fitted. Subsequently, all aspects of the model can be accessed. For example, a simple linear regression via an ordinary least squares would look like the following:
md = smf.ols("ratings ~ category", df_long).fit()
Using the .summary() function we can then get a holistic overview of the model and the outcomes:
print(md.summary())
OLS Regression Results
==============================================================================
Dep. Variable: ratings R-squared: 0.034
Model: OLS Adj. R-squared: 0.031
Method: Least Squares F-statistic: 10.07
Date: Thu, 03 Feb 2022 Prob (F-statistic): 5.05e-05
Time: 15:15:56 Log-Likelihood: -1092.4
No. Observations: 570 AIC: 2191.
Df Residuals: 567 BIC: 2204.
Df Model: 2
Covariance Type: nonrobust
=====================================================================================
coef std err t P>|t| [0.025 0.975]
-------------------------------------------------------------------------------------
Intercept 5.7011 0.174 32.797 0.000 5.360 6.043
category[T.movie] -0.8950 0.202 -4.426 0.000 -1.292 -0.498
category[T.snack] -0.5573 0.206 -2.710 0.007 -0.961 -0.153
==============================================================================
Omnibus: 36.334 Durbin-Watson: 1.597
Prob(Omnibus): 0.000 Jarque-Bera (JB): 40.166
Skew: -0.627 Prob(JB): 1.90e-09
Kurtosis: 2.655 Cond. No. 5.33
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
All displayed information is furthermore directly accessible:
md.bic
2203.869328217725
md.fvalue
10.068893120110223
md.resid
0 -3.806118
1 -3.806118
2 -3.806118
3 -3.806118
4 -3.806118
...
565 1.198889
566 -1.701111
567 -0.581111
568 -0.621111
569 -0.341111
Length: 570, dtype: float64
If we also want to define and run a linear mixed effects model, we only need to change the model type to mixedlm and define our model accordingly:
md = smf.mixedlm("ratings ~ category", df_long, groups=df_long["participant"])
mdf = md.fit(method=["lbfgs"])
print(mdf.summary())
Mixed Linear Model Regression Results
==========================================================================
Model: MixedLM Dependent Variable: ratings
No. Observations: 570 Method: REML
No. Groups: 15 Scale: 2.4044
Min. group size: 38 Log-Likelihood: inf
Max. group size: 38 Converged: Yes
Mean group size: 38.0
--------------------------------------------------------------------------
Coef. Std.Err. z P>|z| [0.025 0.975]
--------------------------------------------------------------------------
Intercept 0.000 3082015.676 0.000 1.000 -6040639.725 6040639.725
category[T.movie] -0.895 0.190 -4.708 0.000 -1.268 -0.522
category[T.snack] -0.557 0.193 -2.882 0.004 -0.936 -0.178
Group Var 0.000
==========================================================================
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/statsmodels/regression/mixed_linear_model.py:1634: UserWarning: Random effects covariance is singular
warnings.warn(msg)
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/statsmodels/regression/mixed_linear_model.py:2054: UserWarning: The random effects covariance matrix is singular.
warnings.warn(_warn_cov_sing)
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/statsmodels/regression/mixed_linear_model.py:2237: ConvergenceWarning: The MLE may be on the boundary of the parameter space.
warnings.warn(msg, ConvergenceWarning)
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/statsmodels/regression/mixed_linear_model.py:2245: UserWarning: The random effects covariance matrix is singular.
warnings.warn(_warn_cov_sing)
/Users/peerherholz/anaconda3/envs/pfp_2021/lib/python3.9/site-packages/statsmodels/regression/mixed_linear_model.py:2261: ConvergenceWarning: The Hessian matrix at the estimated parameter values is not positive definite.
warnings.warn(msg, ConvergenceWarning)
Overall, there’s only one thing left to say regarding statistical analyses in python:

Outro/Q&A - visualization & statistics¶
What we went through in this session was intended as a super small showcase of visualizing and analyzing data via python, specifically using a small subset of available modules which we only just started to explore and have way more functionality.
Sure: this was a very specific use case and data but the steps and underlying principles are transferable to the majority of comparable problems/tasks you might encounter. Especially because the modules we checked are commonly used and are usually the “go-to” option, at least as a first step. As always: make sure to check the fantastic docs of the python modules you’re using, as well as all the fantastic tutorials out there.
Outro/Q&A - working with data in python¶
As mentioned in the beginning, there are so many options of working with data and setting up correspondingly workflows that vary as a function of data at hand and planned analyses. Finding the “only right option” might also not be possible in some cases.
The idea of this section of the course was to outline an example workflow based on some “real-world data” to showcase you some of the capabilities and things you can do using python. Specifically, once more making the case that python has module/library for basically everything you want to do, which are additionally rather straightforward and easy to use and come with comprehensive documentation, as well as many tutorials. This holds true for both domain-general and highly specialized use cases. From data exploration over visualization to analyses you will find a great set of resources that integrates amazingly well with one another and allow you to achieve awesome things in an intelligible and reproducible manner!
The core Python “data science” stack¶
The Python ecosystem contains tens of thousands of packages
Several are very widely used in data science applications:
Jupyter: interactive notebooks
Numpy: numerical computing in Python
pandas: data structures for Python
Scipy: scientific Python tools
Matplotlib: plotting in Python
scikit-learn: machine learning in Python
We’ll cover the first three very briefly here
Other tutorials will go into greater detail on most of the others
The core “Python for psychology” stack¶
The
Python ecosystemcontains tens of thousands ofpackagesSeveral are very widely used in psychology research:
Jupyter: interactive notebooks
Numpy: numerical computing in
Pythonpandas: data structures for
PythonScipy: scientific
PythontoolsMatplotlib: plotting in
Pythonseaborn: plotting in
Pythonscikit-learn: machine learning in
Pythonstatsmodels: statistical analyses in
Pythonpingouin: statistical analyses in
Pythonpsychopy: running experiments in
Pythonnilearn: brain imaging analyses in
Pythonmne: electrophysiology analyses in
Python
Execept
scikit-learn,nilearnandmne, we’ll cover all very briefly in this coursethere are many free tutorials online that will go into greater detail and also cover the other
packages
Homework assignment¶
Your ninth homework assignment will entail working through a few tasks covering the contents discussed in this session within of a jupyter notebook. You can download it here. In order to open it, put the homework assignment notebook within the folder you stored the course materials, start a jupyter notebook as during the sessions, navigate to the homework assignment notebook, open it and have fun!
Deadline: 16/02/2022, 11:59 PM EST
