Overview¶
Here we want to present a brief overview of the dataset and applied analyses before going into detail for certain aspects. Thus we will provide some information regarding the different analyses steps and approaches.
In order to investigate the proposed functional principles of the human auditory cortex, we applied a set of complementary approaches
to versatile dataset. In more detail, we used a recently introduced dataset :cite:`whitehead_singing_2018` within which the auditory categories music,
singing and speech were presented to participants in a passive listening paradigm (while their were watching nature scenes). This stimuli set had the advantage of entailing a progession of spetrotemporal features from fast temporal/low spectral (speech) to intermediate (singing) to slow temporal/broad spectral (music) expression, while their underlying acoustic features were controlled and highly comparable between the categories :cite:`whitehead_singing_2018`. fMRI data was acquired across 2 runs of roughly 8 min each using a 3T [Siemens TIM Trio] scanner running a whole brain multiband accelerated pulse sequence (factor 12) data acquisition protocol :cite:`setsompop_improving_2012` (for further details please see our publication {cite}). The data was then organized according to the Brain Imaging Data Structure (BIDS) :cite:`gorgolewski_brain_2016` and structural images defaced using pydeface :cite:`noauthor_poldracklabpydeface_2020`. Subsequently, the data was quality controlled via MRIQC :cite:`esteban_mriqc_2017` and preprocessed. The strucutral preprocessing was conducted via the BIDS app Mindboggle :cite:`klein_mindboggling_2017` which combines the processing pipelines recon-all from FreeSurfer and antsCorticalThickness from ANTs to extract a multitude of shape features (through the mindboggle pipeline). Notably, we only used the recon-all outputs here while the other derivatives were utilized in a different endeavor. The preprocessing of the functional images entailed a comprehensive nipype workflow consisting of functions from multiple software packages and covered “classic preprocessing” and the generation of cortical gray matter mask. The respective outputs were used within a statistical analyses at the individual level within which we estimated responses to the different sound categories via a GLM, obtaining contrast images. After computing a registration between a given participants native functional space (where we conducted the statistical analyses) and a template/reference space (MNI152 template in its non linear symmetric version), we transformed the the contrast images to this template/reference space. Comparable to the preprocessing, the last three parts were also implemented in nipype. To summarize, here’s a list of the steps:
data conversion to
BIDSdefacing using
pydefacequality control using
MRIQCStrucutral preprocessing using
MindboggleFunctional preprocessing using a
nipype pipelineindividual level statistics using a
nipype pipelineregistration of native functional and template space using a
nipype pipelinetransformation of contrast images from native functional to template space using a
nipype pipeline
The preprocessing, statistical analyses, registration and transformation maps is further explained and accompanied by the corresponding code in the section “Preprocessing & individual level statistics”.
The conducted analyses¶
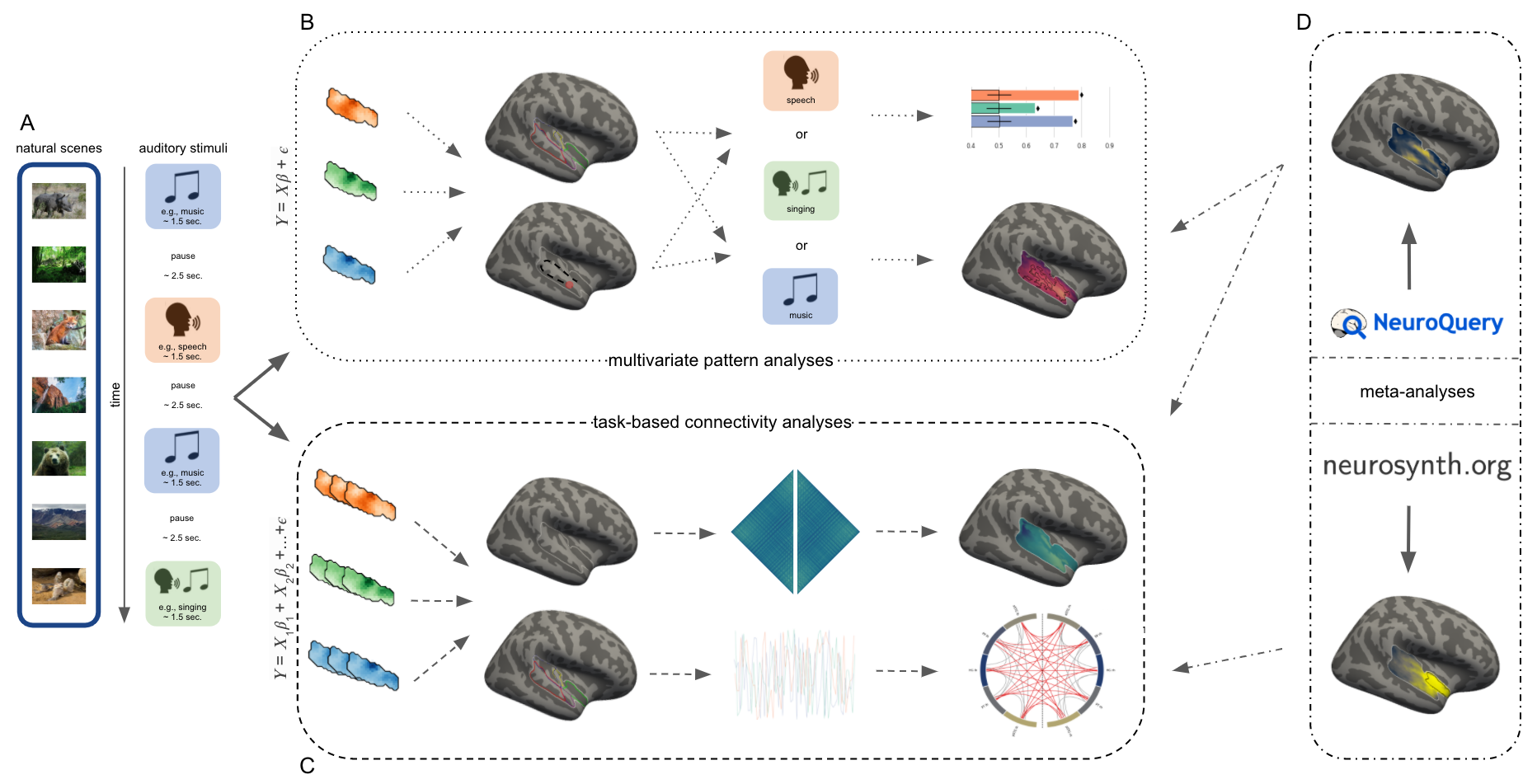
We then applied multivariate (MVPA) and connectivity approaches to investigate spatial and temporal correlates of auditory cortex functional principles respectively. Within both, we evaluated different spatial scales, that is profiles at the voxel and region level. The approaches are summarized in the above presented graphic (Figure 2). Applied to MVPA this refers to running three binary (category pairs) and one multiclass (one vs. rest) classification tasks as searchlight analyses (voxel level) and ROI decoding (region level) (Figure 2 B). Within the context of connectivity this lead to category specific ROI-ROI correlations (region level) and voxel by voxel correlations followed by diffusion map embedding (voxel level) (Figure 2 C). Additionally, we applied meta-analytic approaches to investigate the functional principles across a tremendous amount of research work and to further inform our obtained results, especially category specificity and sensitivity (Figure 2 D). The respective analyses are described in detail, including used code, in the following sections:
9. MVPA
10. Connectivity
11. Meta-analyses
The outcomes¶
We tried to make as many of our outcomes available as possible. This does unfortunately not include the raw data as no corresponding consent was obtained from the participants. Thus, also the steps 1-9 cannot be rerun through the here provided resource. However, we included the corresponding code and as much information as we could. As steps 9-11 utilized derivatives, we can share the respective results along with the code. In turn, these analyses can also be rerun through this setup. To increase the FAIR-ness of everything, we included all that could be shared (code, certain derivatives, etc.) in an OSF repository and brain maps also in a neurovault repository. All of this is explained in further detail in the “Access data” section.