Cortical maps
Contents
Cortical maps¶
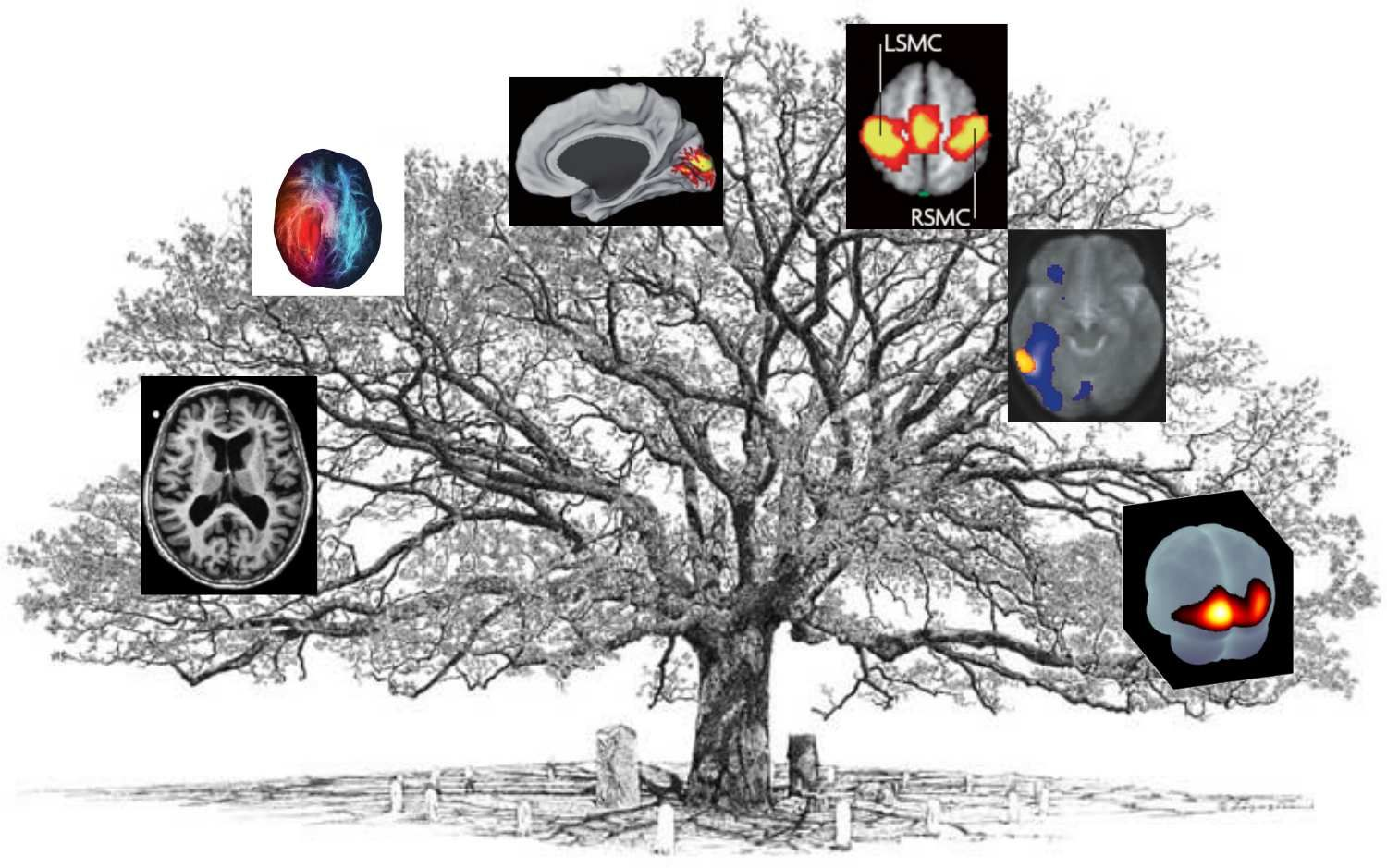
Fig. 1 The neuroimaging tree. Each branch represents one of the techniques that will be briefly presented during this part of the course. Figure adapted by P. Bellec from a variety of non-copyright sources and inspired by the book [Wager and Lindquist, 2015].¶
This first chapter aims to give an overview of this part of the course as a whole. This is a condensed format, which covers all the imaging techniques that we will briefly discuss during this part of the course. If you want to start your project by studying only one chapter, you’ve come to the right place. But this strategy is not recommended! If, on the contrary, you wish to work on the material of each chapter in depth, the essential information can be found elsewhere in the course notes with more details. Nevertheless, this chapter specifies elements of vocabulary, basic notions and will allow you to quickly make connections between the different techniques seen in this part of the course and beyond.
Objectives 📍¶
All of the neuroimaging techniques that we will see have distinct strengths and weaknesses, which make them better suited to different types of application. The general objective of this part of the course is for you to understand if a technique is well suited to a research question. For each technique, we aim more specifically to understand four aspects:
What is the physical principle that allows us to obtain a measurement?
What is the physiological principle, ie what
biological aspectof the brain do we measure?What methods of analysis are needed to be able to interpret the
data?What research questions can be investigated with these
techniques?
Structural and functional imaging¶

Fig. 2 Illustration of the structural and functional techniques briefly discussed in this part of the course, as well as some possible applications in cognitive neuroscience. Figure adapted by P. Bellec from a variety of non-copyright sources and inspired by the book[Wager and Lindquist, 2015].¶
The techniques studied in this part of the course have in common that they aim to generate maps of the brain. They are also central tools in many cognitive neuroscience studies that use neuroimaging. These techniques include:
*Structural Magnetic Resonance Imaging (MRI). This is the best known technique in MRI. It is an image that captures the shape of the brain. It also allows you to see different types of tissue, and in particular the gray matter, that entails the bodies of neurons that are in the brain.
Diffusion MRI (dMRI). This is another type of
imagethat can be acquired with the sameMRI machineasstructural MRI. Thistechniquemakes it possible toreconstructthelarge fiber bundles, i.e. theconnectionsbetweenneurons.Functional MRI (fMRI) is yet another type of
MRI, specifically used to capture and investigatebrain activity. There are two mainanalysis techniquesinfMRI. First,activation mapscan be generated when theparticipantperforms ataskin theMRI. We will thus look for theregionsthat are engaged when theparticipantperforms thistask. Second, analyzes can also be performed when theparticipantsare in aresting state. With that, we will look at theconsistencyofactivitybetweendifferent regions. These arefunctional connectivity cards.Positron Emission Tomography (PET) is a
techniquethat does not useMRI(finally!). Thistechniquerelies onradioactive tracersthat generategamma raysandcamerasthatdetectthesegamma rays. Certaintracers, such asFDG, make it possible to measurecerebral metabolismin relation to theactivityofneurons.Optical imaging measures changes in the
colorofbloodin thebrain, and therefore in itslevelofoxygenation, which is itself linked to theactivityofneurons.
The first two techniques, structural and diffusion MRI, make it possible to study the structure of the brain. The last three techniques (fMRI, PET FDG and optical imaging) all measure functional phenomena. Note that, like MRI, PET can also be used to generate maps of brain structure.
Spatial and temporal resolution¶

Fig. 3 Illustration of the trade-off between temporal and spatial resolution for the neuroimaging techniques briefly discussed in this part of the course. Figure adapted from¶
The techniques seen in this part of the course have in common to have a good spatial resolution, but there are nevertheless significant variations between each of these techniques:
The best from this point of view is the structural MRI whose
spatial resolutionis excellent, withvoxelsof approximately 1 mm\(^3\), that is a cube of1 mm x 1 mm x 1 mm. This allows thestructureof thebrainto be seen in great detail.The dMRI is a little worse, with a
spatial resolutioncloser to2 mm x 2 mm x 2 mm(8 mm\(^3\)).fMRI, on the other hand, commonly uses a
resolutionof3 mm x 3 mm x 3 mm- or 27 mm\(^3\), which is almost 30 times larger than thevoxelof thestructural MRI!Finally, PET and optical imaging have a coarser
spatial resolution, rather equivalent to1 cm x 1 cm x 1 cm(i.e. 1000 mm\(^3\)!!). Even if thePET voxelsare smaller than 1 cm\(^3\), theimageis “blurred” and it is not possible to distinguish small structures.
More on spatial resolution
The notion of spatial resolution generally refers to the minimum size of an object that can be distinguished in an image. If small objects can be distinguished, the resolution is high. If you can only see large objects, the resolution is low. If we are talking about digital photography, the smallest possible object is a pixel, or one of the small squares that make up the image. For maps of the brain, we speak of a voxel, or 3D volume element.
Spatial resolution is not simply the size of a pixel. Two images with the same pixel (or voxel) size can have a different effective resolution if one of the two images is blurry. On the sharp image, smaller objects can be seen than on the blurred image. The effective resolution of the sharp image is therefore higher than that of the blurred image.
Regarding spatial resolution, structural MRI may appear to be the best technique, but there are many other factors to consider when comparing neuroimaging techniques. Another important factor is the temporal resolution. Structural modalities capture changes that are slow to take place. The shape of the cortex and the fiber bundles fall into place throughout development and aging, and they are quite stable even on the scale of several years. In contrast, functional MRI, PET (using FDG), and optical imaging examine brain activity. They measure changes that can occur on the scale of minutes, seconds or even milliseconds.
Temporal resolution
The notion of temporal resolution generally refers to the minimum duration of an event that can be distinguished in a temporal signal. The signals that we see in the course are composed of measurements repeated over time with an interval \(\Delta_t\), generally measured in seconds. We sometimes talk about the sampling frequency, \(f=1/\Delta_t\), measured in Hz. The sampling frequency (Hz) represents the number of measurement points per second.
The temporal resolution does not simply correspond to the time that elapses between two successive measurements \(\Delta_t\). This concept is more difficult to visualize than effective spatial resolution, but is important especially in the case of optical imaging. Optical imaging captures a slow vascular phenomenon. So even if we have peaks of activity separated in time at the neuronal level, if the time interval between the peaks is too short we will only see a single event at the vascular level. It is the equivalent of a blurred image, but in the temporal dimension.
Magnetic resonance imaging¶
Fig. 4 A functional magnetic resonance imaging machine. Image shutterstock ID 1866109303.¶
An MRI machine is an imposing machine that can weigh several tens of tons! The most obvious element in an MRI system is the relatively deep tunnel, which is a giant magnet. The participant is positioned on a table which can move to bring the participant to the center of the magnet. The reason for placing the research participant there is that the magnetic field in the center of the magnet is very homogeneous and points in a constant direction. This homogeneous magnetic field, called B0, is like a blank canvas for a painting. Smaller magnets, called gradients, will be turned on and then off quickly to modify the magnetic field in different parts of the brain. As different biological tissues react differently to these stimuli, the gradients allow us to “paint” a picture of the brain on the “canvas” B0. We actually acquire a series of images that will form a 3D volume covering the entire brain. We will discuss the physical process of generating an MRI image in more detail in chapter Functional MRI.
Structural MRI¶
%matplotlib inline
# This code retrieves T1 MRI data
# and generates an image in three planes of cuts
# ignore warnings
import warnings
warnings.filterwarnings("ignore")
# Download an anatomical scan (here the MNI152 template)
from nilearn.datasets import fetch_icbm152_2009
mni = fetch_icbm152_2009()
# Visualize the 3D brain volume
import matplotlib.pyplot as plt
from myst_nb import glue
from nilearn.plotting import plot_anat, view_img
t1_fig = plt.figure(figsize=(12, 4))
plot_anat(
mni.t1,
axes=t1_fig.gca(),
black_bg=False,
dim=0,
cut_coords=[-17, 0, 17],
title='T1 weighted MRI',
output_file='../../../static/neuroscience/mri_example.png'
)
t1_fig_int = view_img(
mni.t1,
cmap='bone',
colorbar=False,
bg_img=False,
dim=-2,
symmetric_cmap=False,
title='T1 weighted MRI'
)
glue("t1_fig_int", t1_fig_int, display=False)
