Iterables¶
Some steps in a neuroimaging analysis are repetitive. Running the same preprocessing on multiple subjects or doing statistical inference on multiple files. To prevent the creation of multiple individual scripts, Nipype has as execution plugin for Workflow, called iterables.

If you are interested in more advanced procedures, such as synchronizing multiple iterables or using conditional iterables, check out the synchronize and intersource section in the JoinNode notebook.
Realistic example¶
Let’s assume we have a workflow with two nodes, node (A) does simple skull stripping, and is followed by a node (B) that does isometric smoothing. Now, let’s say, that we are curious about the effect of different smoothing kernels. Therefore, we want to run the smoothing node with FWHM set to 2mm, 8mm, and 16mm.
from nipype import Node, Workflow
from nipype.interfaces.fsl import BET, IsotropicSmooth
# Initiate a skull stripping Node with BET
skullstrip = Node(BET(mask=True,
in_file='/data/ds000114/sub-01/ses-test/anat/sub-01_ses-test_T1w.nii.gz'),
name="skullstrip")
Create a smoothing Node with IsotropicSmooth
isosmooth = Node(IsotropicSmooth(), name='iso_smooth')
Now, to use iterables and therefore smooth with different fwhm is as simple as that:
isosmooth.iterables = ("fwhm", [4, 8, 16])
And to wrap it up. We need to create a workflow, connect the nodes and finally, can run the workflow in parallel.
# Create the workflow
wf = Workflow(name="smoothflow")
wf.base_dir = "/output"
wf.connect(skullstrip, 'out_file', isosmooth, 'in_file')
# Run it in parallel (one core for each smoothing kernel)
wf.run('MultiProc', plugin_args={'n_procs': 3})
211017-17:19:31,855 nipype.workflow INFO:
Workflow smoothflow settings: ['check', 'execution', 'logging', 'monitoring']
211017-17:19:31,860 nipype.workflow INFO:
Running in parallel.
211017-17:19:31,864 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 3/3.
211017-17:19:31,922 nipype.workflow INFO:
[Node] Setting-up "smoothflow.skullstrip" in "/output/smoothflow/skullstrip".
211017-17:19:31,933 nipype.workflow INFO:
[Node] Running "skullstrip" ("nipype.interfaces.fsl.preprocess.BET"), a CommandLine Interface with command:
bet /data/ds000114/sub-01/ses-test/anat/sub-01_ses-test_T1w.nii.gz /output/smoothflow/skullstrip/sub-01_ses-test_T1w_brain.nii.gz -m
211017-17:19:33,868 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 2/3.
Currently running:
* smoothflow.skullstrip
211017-17:19:35,871 nipype.workflow INFO:
[Node] Finished "smoothflow.skullstrip".
211017-17:19:37,873 nipype.workflow INFO:
[Job 0] Completed (smoothflow.skullstrip).
211017-17:19:37,876 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 3/3.
211017-17:19:37,934 nipype.workflow INFO:
[Node] Setting-up "smoothflow.iso_smooth" in "/output/smoothflow/_fwhm_4/iso_smooth".
211017-17:19:37,935 nipype.workflow INFO:
[Node] Setting-up "smoothflow.iso_smooth" in "/output/smoothflow/_fwhm_8/iso_smooth".
211017-17:19:37,935 nipype.workflow INFO:
[Node] Setting-up "smoothflow.iso_smooth" in "/output/smoothflow/_fwhm_16/iso_smooth".
211017-17:19:37,953 nipype.workflow INFO:
[Node] Running "iso_smooth" ("nipype.interfaces.fsl.maths.IsotropicSmooth"), a CommandLine Interface with command:
fslmaths /output/smoothflow/skullstrip/sub-01_ses-test_T1w_brain.nii.gz -s 1.69864 /output/smoothflow/_fwhm_4/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz
211017-17:19:37,964 nipype.workflow INFO:
[Node] Running "iso_smooth" ("nipype.interfaces.fsl.maths.IsotropicSmooth"), a CommandLine Interface with command:
fslmaths /output/smoothflow/skullstrip/sub-01_ses-test_T1w_brain.nii.gz -s 3.39729 /output/smoothflow/_fwhm_8/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz
211017-17:19:37,964 nipype.workflow INFO:
[Node] Running "iso_smooth" ("nipype.interfaces.fsl.maths.IsotropicSmooth"), a CommandLine Interface with command:
fslmaths /output/smoothflow/skullstrip/sub-01_ses-test_T1w_brain.nii.gz -s 6.79457 /output/smoothflow/_fwhm_16/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz
211017-17:19:39,876 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 1.15/1.75, Free processors: 0/3.
Currently running:
* smoothflow.iso_smooth
* smoothflow.iso_smooth
* smoothflow.iso_smooth
211017-17:19:41,864 nipype.workflow INFO:
[Node] Finished "smoothflow.iso_smooth".
211017-17:19:41,877 nipype.workflow INFO:
[Job 3] Completed (smoothflow.iso_smooth).
211017-17:19:41,880 nipype.workflow INFO:
[MultiProc] Running 2 tasks, and 0 jobs ready. Free memory (GB): 1.35/1.75, Free processors: 1/3.
Currently running:
* smoothflow.iso_smooth
* smoothflow.iso_smooth
211017-17:19:44,557 nipype.workflow INFO:
[Node] Finished "smoothflow.iso_smooth".
211017-17:19:45,881 nipype.workflow INFO:
[Job 2] Completed (smoothflow.iso_smooth).
211017-17:19:45,884 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 2/3.
Currently running:
* smoothflow.iso_smooth
211017-17:19:50,524 nipype.workflow INFO:
[Node] Finished "smoothflow.iso_smooth".
211017-17:19:51,894 nipype.workflow INFO:
[Job 1] Completed (smoothflow.iso_smooth).
211017-17:19:51,899 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 3/3.
<networkx.classes.digraph.DiGraph at 0x7f4d81d00910>
Note, that iterables is set on a specific node (isosmooth in this case), but Workflow is needed to expend the graph to three subgraphs with three different versions of the isosmooth node.
If we visualize the graph with exec, we can see where the parallelization actually takes place.
# Visualize the detailed graph
from IPython.display import Image
wf.write_graph(graph2use='exec', format='png', simple_form=True)
Image(filename='/output/smoothflow/graph_detailed.png')
211017-17:19:54,181 nipype.workflow INFO:
Generated workflow graph: /output/smoothflow/graph.png (graph2use=exec, simple_form=True).
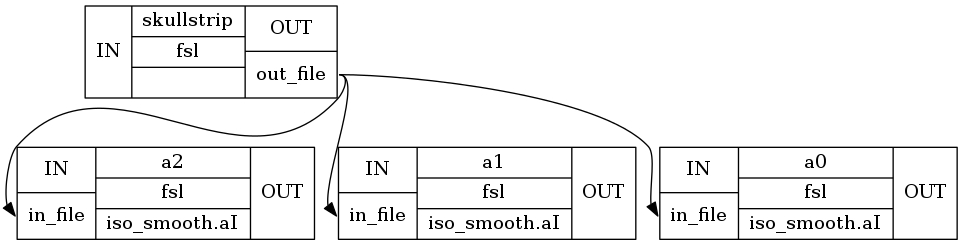
If you look at the structure in the workflow directory, you can also see, that for each smoothing, a specific folder was created, i.e. _fwhm_16.
!tree /output/smoothflow -I '*txt|*pklz|report*|*.json|*js|*.dot|*.html'
/output/smoothflow
├── _fwhm_16
│ └── iso_smooth
│ ├── _report
│ └── sub-01_ses-test_T1w_brain_smooth.nii.gz
├── _fwhm_4
│ └── iso_smooth
│ ├── _report
│ └── sub-01_ses-test_T1w_brain_smooth.nii.gz
├── _fwhm_8
│ └── iso_smooth
│ ├── _report
│ └── sub-01_ses-test_T1w_brain_smooth.nii.gz
├── graph_detailed.png
├── graph.png
└── skullstrip
├── _report
└── sub-01_ses-test_T1w_brain.nii.gz
11 directories, 6 files
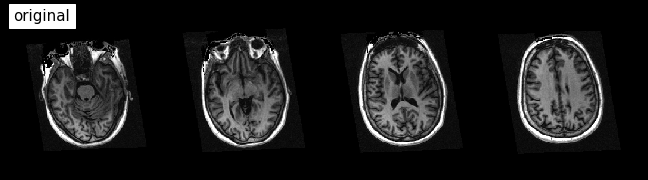
Now, let’s visualize the results!
from nilearn import plotting
%matplotlib inline
plotting.plot_anat(
'/data/ds000114/sub-01/ses-test/anat/sub-01_ses-test_T1w.nii.gz', title='original',
display_mode='z', dim=-1, cut_coords=(-50, -35, -20, -5), annotate=False);
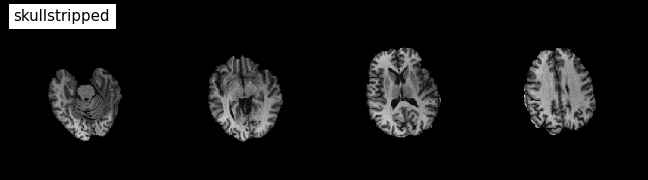
plotting.plot_anat(
'/output/smoothflow/skullstrip/sub-01_ses-test_T1w_brain.nii.gz', title='skullstripped',
display_mode='z', dim=-1, cut_coords=(-50, -35, -20, -5), annotate=False);
plotting.plot_anat(
'/output/smoothflow/_fwhm_4/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz', title='FWHM=4',
display_mode='z', dim=-0.5, cut_coords=(-50, -35, -20, -5), annotate=False);
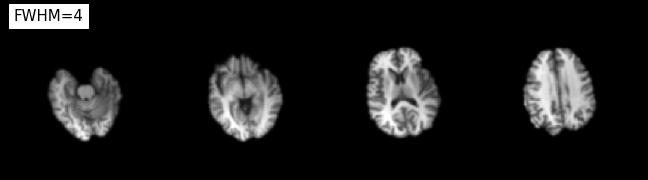
plotting.plot_anat(
'/output/smoothflow/_fwhm_8/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz', title='FWHM=8',
display_mode='z', dim=-0.5, cut_coords=(-50, -35, -20, -5), annotate=False);
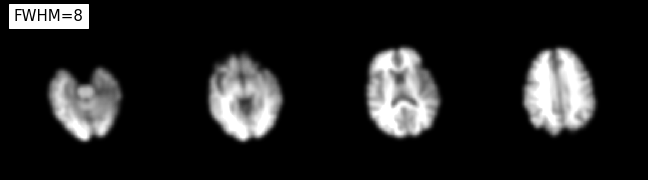
plotting.plot_anat(
'/output/smoothflow/_fwhm_16/iso_smooth/sub-01_ses-test_T1w_brain_smooth.nii.gz', title='FWHM=16',
display_mode='z', dim=-0.5, cut_coords=(-50, -35, -20, -5), annotate=False);

IdentityInterface (special use case of iterables)¶
We often want to start our worflow from creating subgraphs, e.g. for running preprocessing for all subjects. We can easily do it with setting iterables on the IdentityInterface. The IdentityInterface interface allows you to create Nodes that does simple identity mapping, i.e. Nodes that only work on parameters/strings.
For example, you want to start your workflow by collecting anatomical files for 5 subjects.
# First, let's specify the list of subjects
subject_list = ['01', '02', '03', '07']
Now, we can create the IdentityInterface Node
from nipype import IdentityInterface
infosource = Node(IdentityInterface(fields=['subject_id']),
name="infosource")
infosource.iterables = [('subject_id', subject_list)]
That’s it. Now, we can connect the output fields of this infosource node to SelectFiles and DataSink nodes.
from os.path import join as opj
from nipype.interfaces.io import SelectFiles, DataSink
anat_file = opj('sub-{subject_id}', 'ses-test', 'anat', 'sub-{subject_id}_ses-test_T1w.nii.gz')
templates = {'anat': anat_file}
selectfiles = Node(SelectFiles(templates,
base_directory='/data/ds000114'),
name="selectfiles")
# Datasink - creates output folder for important outputs
datasink = Node(DataSink(base_directory="/output",
container="datasink"),
name="datasink")
wf_sub = Workflow(name="choosing_subjects")
wf_sub.connect(infosource, "subject_id", selectfiles, "subject_id")
wf_sub.connect(selectfiles, "anat", datasink, "anat_files")
wf_sub.run()
211017-17:20:22,683 nipype.workflow INFO:
Workflow choosing_subjects settings: ['check', 'execution', 'logging', 'monitoring']
211017-17:20:22,748 nipype.workflow INFO:
Running serially.
211017-17:20:22,749 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.selectfiles" in "/tmp/tmpk5q8x_b2/choosing_subjects/_subject_id_07/selectfiles".
211017-17:20:22,757 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211017-17:20:22,780 nipype.workflow INFO:
[Node] Finished "choosing_subjects.selectfiles".
211017-17:20:22,782 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.datasink" in "/tmp/tmpy6rsnaja/choosing_subjects/_subject_id_07/datasink".
211017-17:20:22,799 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211017-17:20:22,841 nipype.workflow INFO:
[Node] Finished "choosing_subjects.datasink".
211017-17:20:22,842 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.selectfiles" in "/tmp/tmpfornacdh/choosing_subjects/_subject_id_03/selectfiles".
211017-17:20:22,849 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211017-17:20:22,866 nipype.workflow INFO:
[Node] Finished "choosing_subjects.selectfiles".
211017-17:20:22,868 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.datasink" in "/tmp/tmp5a4hkm9b/choosing_subjects/_subject_id_03/datasink".
211017-17:20:22,885 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211017-17:20:22,914 nipype.workflow INFO:
[Node] Finished "choosing_subjects.datasink".
211017-17:20:22,917 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.selectfiles" in "/tmp/tmp210f17gd/choosing_subjects/_subject_id_02/selectfiles".
211017-17:20:22,923 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211017-17:20:22,936 nipype.workflow INFO:
[Node] Finished "choosing_subjects.selectfiles".
211017-17:20:22,939 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.datasink" in "/tmp/tmpudpvc77z/choosing_subjects/_subject_id_02/datasink".
211017-17:20:22,970 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211017-17:20:23,82 nipype.workflow INFO:
[Node] Finished "choosing_subjects.datasink".
211017-17:20:23,89 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.selectfiles" in "/tmp/tmpdp0gzp83/choosing_subjects/_subject_id_01/selectfiles".
211017-17:20:23,110 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211017-17:20:23,152 nipype.workflow INFO:
[Node] Finished "choosing_subjects.selectfiles".
211017-17:20:23,155 nipype.workflow INFO:
[Node] Setting-up "choosing_subjects.datasink" in "/tmp/tmptj1wa1op/choosing_subjects/_subject_id_01/datasink".
211017-17:20:23,172 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211017-17:20:23,204 nipype.workflow INFO:
[Node] Finished "choosing_subjects.datasink".
<networkx.classes.digraph.DiGraph at 0x7f4d75319d10>
Now we can check that five anatomicl images are in anat_files directory:
! ls -lh /output/datasink/anat_files/
total 34M
-rwxr--r-- 2 neuro root 8.3M Nov 5 2020 sub-01_ses-test_T1w.nii.gz
-rwxr--r-- 2 neuro root 9.6M Nov 5 2020 sub-02_ses-test_T1w.nii.gz
-rwxr--r-- 2 neuro root 7.7M Nov 5 2020 sub-03_ses-test_T1w.nii.gz
-rwxr--r-- 2 neuro root 8.2M Nov 5 2020 sub-07_ses-test_T1w.nii.gz
This was just a simple example of using IdentityInterface, but a complete example of preprocessing workflow you can find in Preprocessing Example).
Exercise 1¶
Create a workflow to calculate various powers of 2 using two nodes, one for IdentityInterface with iterables, and one for Function interface to calculate the power of 2.
# write your solution here
# lets start from the Identity node
from nipype import Function, Node, Workflow
from nipype.interfaces.utility import IdentityInterface
iden = Node(IdentityInterface(fields=['number']), name="identity")
iden.iterables = [("number", range(8))]
# the second node should use the Function interface
def power_of_two(n):
return 2**n
# Create Node
power = Node(Function(input_names=["n"],
output_names=["pow"],
function=power_of_two),
name='power')
#and now the workflow
wf_ex1 = Workflow(name="exercise1")
wf_ex1.connect(iden, "number", power, "n")
res_ex1 = wf_ex1.run()
# we can print the results
for i in range(8):
print(list(res_ex1.nodes())[i].result.outputs)
211017-17:20:24,389 nipype.workflow INFO:
Workflow exercise1 settings: ['check', 'execution', 'logging', 'monitoring']
211017-17:20:24,446 nipype.workflow INFO:
Running serially.
211017-17:20:24,448 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmp7fi3z2u3/exercise1/_number_7/power".
211017-17:20:24,453 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,462 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,464 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmp776t51tz/exercise1/_number_6/power".
211017-17:20:24,471 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,479 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,482 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmpmm9vfjkg/exercise1/_number_5/power".
211017-17:20:24,488 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,496 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,497 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmpi9yx_d7c/exercise1/_number_4/power".
211017-17:20:24,503 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,511 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,513 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmp2w9uvm3p/exercise1/_number_3/power".
211017-17:20:24,521 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,529 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,531 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmp1xts32fw/exercise1/_number_2/power".
211017-17:20:24,536 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,543 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,544 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmpjqxxi2ov/exercise1/_number_1/power".
211017-17:20:24,549 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,555 nipype.workflow INFO:
[Node] Finished "exercise1.power".
211017-17:20:24,557 nipype.workflow INFO:
[Node] Setting-up "exercise1.power" in "/tmp/tmps6e8t0xy/exercise1/_number_0/power".
211017-17:20:24,562 nipype.workflow INFO:
[Node] Running "power" ("nipype.interfaces.utility.wrappers.Function")
211017-17:20:24,571 nipype.workflow INFO:
[Node] Finished "exercise1.power".
pow = 1
pow = 2
pow = 4
pow = 8
pow = 16
pow = 32
pow = 64
pow = 128