Introduction to Statistics in python¶
In this section, we will cover how you can use Python to do some statistics. There are many packages to do so, but we will focus on four:
This notebook is strongly based on the scipy-lectures.org section about statistics.
Data representation and interaction¶
Data as a table¶
The setting that we consider for statistical analysis is that of multiple observations or samples described by a set of different attributes or features. The data can than be seen as a 2D table, or matrix, with columns giving the different attributes of the data, and rows the observations. For example purposes, we will make use of phenotypic information within a dataset from OpenNeuro, provided as part of the following publication:
It contains sex, age, handedness, group id, Mood and Feelings Questionnaire total scores and Snaith-Hamilton Pleasure Scale total scores for a fair amount of participants.
We will use curl to gather the respective file:
%%bash
curl https://openneuro.org/crn/datasets/ds003709/snapshots/1.0.0/files/participants.tsv -o /data/participants_stats.tsv
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
100 1761 0 1761 0 0 14238 0 --:--:-- --:--:-- --:--:-- 14201
!head /data/participants_stats.tsv
participant_id sex age handedness group MFQ SHAPS
sub-20900 M 17 RIGHT HV 0.0 1.0
sub-22686 M 16 RIGHT HV 2.0 10.0
sub-23017 F 17 RIGHT MDD 15.0 6.0
sub-23108 F 16 RIGHT HV 0.0 7.0
sub-23303 F 16 RIGHT HV 0.0 0.0
sub-23399 F 17 RIGHT MDD 1.0 14.0
sub-23520 F 17 RIGHT MDD 7.0 22.0
sub-23540 F 16 RIGHT MDD 5.0 21.0
sub-23546 F 17 RIGHT MDD 1.0 0.0

The pandas data-frame¶
Creating dataframes: reading data files or converting arrays¶
Reading from a CSV file¶
Using the above CSV file that gives observations of brain size and weight and IQ (Willerman et al. 1991), the data are a mixture of numerical and categorical values::
import pandas as pd
data = pd.read_csv('/data/participants_stats.tsv', sep='\t', na_values=".")
data
| participant_id | sex | age | handedness | group | MFQ | SHAPS | |
|---|---|---|---|---|---|---|---|
| 0 | sub-20900 | M | 17 | RIGHT | HV | 0.0 | 1.0 |
| 1 | sub-22686 | M | 16 | RIGHT | HV | 2.0 | 10.0 |
| 2 | sub-23017 | F | 17 | RIGHT | MDD | 15.0 | 6.0 |
| 3 | sub-23108 | F | 16 | RIGHT | HV | 0.0 | 7.0 |
| 4 | sub-23303 | F | 16 | RIGHT | HV | 0.0 | 0.0 |
| 5 | sub-23399 | F | 17 | RIGHT | MDD | 1.0 | 14.0 |
| 6 | sub-23520 | F | 17 | RIGHT | MDD | 7.0 | 22.0 |
| 7 | sub-23540 | F | 16 | RIGHT | MDD | 5.0 | 21.0 |
| 8 | sub-23546 | F | 17 | RIGHT | MDD | 1.0 | 0.0 |
| 9 | sub-23549 | M | 16 | RIGHT | MDD | 4.0 | 20.0 |
| 10 | sub-23564 | F | 16 | RIGHT | MDD | 2.0 | 15.0 |
| 11 | sub-23576 | F | 15 | RIGHT | MDD | 9.0 | 4.0 |
| 12 | sub-23593 | F | 15 | RIGHT | MDD | 16.0 | 18.0 |
| 13 | sub-23607 | F | 17 | RIGHT | MDD | 6.0 | 2.0 |
| 14 | sub-23613 | F | 14 | RIGHT | HV | NaN | NaN |
| 15 | sub-23625 | F | 14 | RIGHT | MDD | 8.0 | 8.0 |
| 16 | sub-23639 | F | 17 | RIGHT | HV | 4.0 | 1.0 |
| 17 | sub-23643 | F | 17 | RIGHT | MDD | 13.0 | 17.0 |
| 18 | sub-23649 | F | 15 | RIGHT | MDD | 15.0 | 16.0 |
| 19 | sub-23652 | M | 16 | RIGHT | MDD | 8.0 | 17.0 |
| 20 | sub-23673 | F | 17 | RIGHT | HV | 0.0 | 5.0 |
| 21 | sub-23674 | F | 15 | RIGHT | MDD | 6.0 | 9.0 |
| 22 | sub-23689 | F | 18 | RIGHT | MDD | 5.0 | 3.0 |
| 23 | sub-23717 | M | 16 | AMBIDEXTROUS | MDD | 9.0 | 14.0 |
| 24 | sub-23718 | F | 17 | RIGHT | MDD | NaN | NaN |
| 25 | sub-23720 | F | 16 | RIGHT | MDD | 5.0 | 12.0 |
| 26 | sub-23757 | F | 14 | RIGHT | MDD | 1.0 | 1.0 |
| 27 | sub-23780 | F | 18 | RIGHT | MDD | 15.0 | 3.0 |
| 28 | sub-23783 | F | 14 | LEFT | MDD | NaN | NaN |
| 29 | sub-23798 | F | 15 | RIGHT | MDD | 8.0 | 8.0 |
| 30 | sub-23814 | F | 13 | RIGHT | MDD | 20.0 | 10.0 |
| 31 | sub-23817 | F | 15 | LEFT | HV | 0.0 | 0.0 |
| 32 | sub-23820 | F | 16 | RIGHT | HV | 0.0 | 0.0 |
| 33 | sub-23823 | M | 17 | RIGHT | MDD | 5.0 | 21.0 |
| 34 | sub-23830 | M | 16 | LEFT | MDD | 14.0 | 26.0 |
| 35 | sub-23834 | F | 13 | RIGHT | HV | 2.0 | 2.0 |
| 36 | sub-23843 | F | 16 | RIGHT | HV | 1.0 | 5.0 |
| 37 | sub-23856 | F | 12 | RIGHT | HV | 1.0 | 14.0 |
| 38 | sub-23866 | M | 16 | RIGHT | HV | 4.0 | 15.0 |
| 39 | sub-23867 | F | 14 | AMBIDEXTROUS | MDD | 11.0 | 25.0 |
| 40 | sub-23880 | F | 17 | RIGHT | HV | 0.0 | 0.0 |
| 41 | sub-23890 | F | 14 | RIGHT | HV | 0.0 | 2.0 |
| 42 | sub-23901 | M | 15 | RIGHT | HV | NaN | 0.0 |
| 43 | sub-23903 | F | 15 | RIGHT | MDD | 16.0 | 11.0 |
| 44 | sub-23905 | F | 16 | RIGHT | HV | 4.0 | 10.0 |
| 45 | sub-23919 | M | 14 | LEFT | MDD | 0.0 | 1.0 |
| 46 | sub-23937 | F | 15 | AMBIDEXTROUS | HV | 0.0 | 0.0 |
| 47 | sub-23943 | F | 16 | RIGHT | HV | 8.0 | NaN |
| 48 | sub-23951 | F | 13 | RIGHT | MDD | 8.0 | 11.0 |
| 49 | sub-23952 | M | 15 | RIGHT | MDD | 5.0 | 16.0 |
| 50 | sub-23969 | F | 15 | RIGHT | HV | NaN | NaN |
Creating from arrays¶
A pandas.DataFrame can also be seen as a dictionary of 1D ‘series’, eg arrays or lists. If we have 3 numpy arrays:
import numpy as np
t = np.linspace(-6, 6, 20)
sin_t = np.sin(t)
cos_t = np.cos(t)
We can expose them as a pandas.DataFrame:
pd.DataFrame({'t': t, 'sin': sin_t, 'cos': cos_t}).head()
| t | sin | cos | |
|---|---|---|---|
| 0 | -6.000000 | 0.279415 | 0.960170 |
| 1 | -5.368421 | 0.792419 | 0.609977 |
| 2 | -4.736842 | 0.999701 | 0.024451 |
| 3 | -4.105263 | 0.821291 | -0.570509 |
| 4 | -3.473684 | 0.326021 | -0.945363 |
Other inputs: pandas can input data from SQL, excel files, or other formats. See the pandas documentation.
Manipulating data¶
data is a pandas.DataFrame, that resembles R’s dataframe:
data.shape # 51 rows and 7 columns
(51, 7)
data.columns # It has columns
Index(['participant_id', 'sex', 'age', 'handedness', 'group', 'MFQ', 'SHAPS'], dtype='object')
print(data['group'].head()) # Columns can be addressed by name
0 HV
1 HV
2 MDD
3 HV
4 HV
Name: group, dtype: object
# Simpler selector
data[data['group'] == 'HV']['MFQ'].mean()
1.5294117647058822
Note: For a quick view on a large dataframe, use its describe pandas.DataFrame.describe.
data.describe()
| age | MFQ | SHAPS | |
|---|---|---|---|
| count | 51.000000 | 46.000000 | 46.000000 |
| mean | 15.568627 | 5.739130 | 9.195652 |
| std | 1.360219 | 5.511341 | 7.759208 |
| min | 12.000000 | 0.000000 | 0.000000 |
| 25% | 15.000000 | 1.000000 | 2.000000 |
| 50% | 16.000000 | 5.000000 | 8.500000 |
| 75% | 17.000000 | 8.000000 | 15.000000 |
| max | 18.000000 | 20.000000 | 26.000000 |
# Frequency count for a given column
data['age'].value_counts()
16 15
17 12
15 11
14 7
13 3
18 2
12 1
Name: age, dtype: int64
# Dummy-code # of group (i.e., get N-binary columns)
pd.get_dummies(data['group'])[:15]
| HV | MDD | |
|---|---|---|
| 0 | 1 | 0 |
| 1 | 1 | 0 |
| 2 | 0 | 1 |
| 3 | 1 | 0 |
| 4 | 1 | 0 |
| 5 | 0 | 1 |
| 6 | 0 | 1 |
| 7 | 0 | 1 |
| 8 | 0 | 1 |
| 9 | 0 | 1 |
| 10 | 0 | 1 |
| 11 | 0 | 1 |
| 12 | 0 | 1 |
| 13 | 0 | 1 |
| 14 | 1 | 0 |
The split-apply-combine pattern¶
A very common data processing strategy is to…
Split the dataset into groups
Apply some operation(s) to each group
(Optionally) combine back into one dataset
Pandas provides powerful and fast tools for this. For example the groupby function.
groupby: splitting a dataframe on values of categorical variables:
groupby_group = data.groupby('group')
for group, value in groupby_group['MFQ']:
print((group, value.mean()))
('HV', 1.5294117647058822)
('MDD', 8.206896551724139)
groupby_group is a powerful object that exposes many operations on the resulting group of dataframes:
groupby_group.mean()
| age | MFQ | SHAPS | |
|---|---|---|---|
| group | |||
| HV | 15.450000 | 1.529412 | 4.235294 |
| MDD | 15.645161 | 8.206897 | 12.103448 |
Plotting data¶
Pandas comes with some plotting tools (pandas.tools.plotting, using
matplotlib behind the scene) to display statistics of the data in
dataframes.
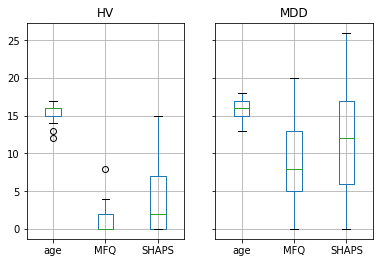
For example, let’s use boxplot (in this case even groupby_group.boxplot) to better understand the structure of the data.
%matplotlib inline
groupby_group.boxplot(column=['age', 'MFQ', 'SHAPS']);
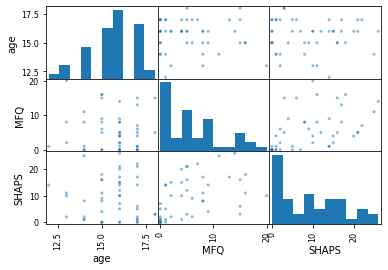
Scatter matrices¶
pd.plotting.scatter_matrix(data[['age', 'MFQ', 'SHAPS']]);
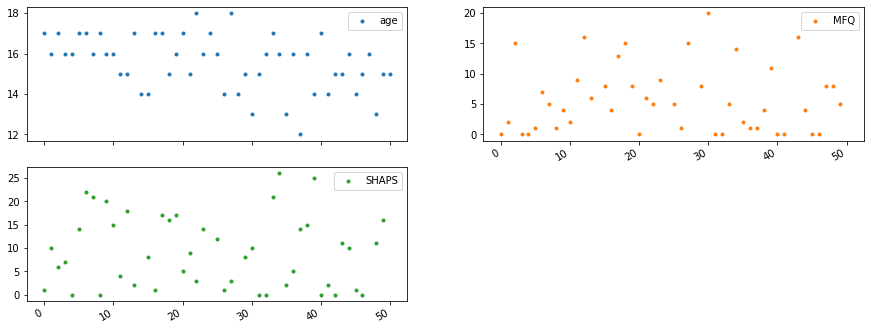
Plot content of a dataset¶
data.head()
| participant_id | sex | age | handedness | group | MFQ | SHAPS | |
|---|---|---|---|---|---|---|---|
| 0 | sub-20900 | M | 17 | RIGHT | HV | 0.0 | 1.0 |
| 1 | sub-22686 | M | 16 | RIGHT | HV | 2.0 | 10.0 |
| 2 | sub-23017 | F | 17 | RIGHT | MDD | 15.0 | 6.0 |
| 3 | sub-23108 | F | 16 | RIGHT | HV | 0.0 | 7.0 |
| 4 | sub-23303 | F | 16 | RIGHT | HV | 0.0 | 0.0 |
data.plot(lw=0, marker='.', subplots=True, layout=(-1, 2), figsize=(15, 6));

Pingouin is an open-source statistical package written in Python 3 and based mostly on Pandas and NumPy.¶
ANOVAs: one- and two-ways, repeated measures, mixed, ancova
Post-hocs tests and pairwise comparisons
Robust correlations
Partial correlation, repeated measures correlation and intraclass correlation
Linear/logistic regression and mediation analysis
Bayesian T-test and Pearson correlation
Tests for sphericity, normality and homoscedasticity
Effect sizes and power analysis
Parametric/bootstrapped confidence intervals around an effect size or a correlation coefficient
Circular statistics
Plotting: Bland-Altman plot, Q-Q plot, etc…
Pingouin is designed for users who want simple yet exhaustive statistical functions.
**material scavenged from 10 minutes to Pingouin and the pingouin docs¶
import pingouin as pg
“In the broadest sense correlation is any statistical association, though in common usage it most often refers to how close two variables are to having a linear relationship with each other” - Wikipedia
When talking about correlation, we commonly mean the Pearson correlation coefficient, also referred to as Pearson’s r

pearson_correlation = pg.corr(data['MFQ'], data['SHAPS'])
display(pearson_correlation)
cor_coeeficient = pearson_correlation['r']
n = len(data) # sample size
| n | r | CI95% | r2 | adj_r2 | p-val | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| pearson | 45 | 0.457 | [0.19, 0.66] | 0.209 | 0.171 | 0.001601 | 16.321 | 0.898 |
Test summary¶
‘n’ : Sample size (after NaN removal)
‘outliers’ : number of outliers (only for ‘shepherd’ or ‘skipped’)
‘r’ : Correlation coefficient
‘CI95’ : 95% parametric confidence intervals
‘r2’ : R-squared
‘adj_r2’ : Adjusted R-squared
‘p-val’ : one or two tailed p-value
‘BF10’ : Bayes Factor of the alternative hypothesis (Pearson only)
‘power’ : achieved power of the test (= 1 - type II error)
Before we calculate: Testing statistical premises¶
Statistical procedures can be classfied into either parametric or non parametric prcedures, which require different necessary preconditions to be met in order to show consistent/robust results.
Generally people assume that their data follows a gaussian distribution, which allows for parametric tests to be run.
Nevertheless it is essential to first test the distribution of your data to decide if the assumption of normally distributed data holds, if this is not the case we would have to switch to non parametric tests.
Shapiro Wilk normality test¶
Standard procedure to test for normal distribution. Tests if the distribution of you data deviates significtanly from a normal distribution. returns:
normal:booleanTrueifxcomes from anormal distribution.p:floatP-value.
# Return a boolean (true if normal) and the associated p-value
pg.normality(data['MFQ'])
(True, 1.0)
Henze-Zirkler multivariate normality test¶
Same procedure for multivariate normal distributions
returns
normal : boolean True if X comes from a multivariate normal distribution.
p : float P-value.
# Return a boolean (true if normal) and the associated p-value
import numpy as np
np.random.seed(123)
mean, cov, n = [4, 6], [[1, .5], [.5, 1]], 30
X = np.random.multivariate_normal(mean, cov, n)
pg.multivariate_normality(X, alpha=.05)
(True, 0.7523511059223016)
Mauchly test for sphericity¶
“Sphericity is the condition where the variances of the differences between all combinations of related groups (levels) are equal. Violation of sphericity is when the variances of the differences between all combinations of related groups are not equal.” - https://statistics.laerd.com/statistical-guides/sphericity-statistical-guide.php
returns
spher : boolean True if data have the sphericity property.
W : float Test statistic
chi_sq : float Chi-square statistic
ddof : int Degrees of freedom
p : float P-value.
pg.sphericity(data)
(False, 0.002, 302.823, 20, 2.142051582323624e-52)
Testing for homoscedasticity¶
“In statistics, a sequence or a vector of random variables is homoscedastic /ˌhoʊmoʊskəˈdæstɪk/ if all random variables in the sequence or vector have the same finite variance.” - wikipedia
returns:
equal_var : boolean True if data have equal variance.
p : float P-value.
Note: This function first tests if the data are normally distributed using the Shapiro-Wilk test. If yes, then the homogeneity of variances is measured using the Bartlett test. If the data are not normally distributed, the Levene test, which is less sensitive to departure from normality, is used.
np.random.seed(123)
# Scale = standard deviation of the distribution.
array_1 = np.random.normal(loc=0, scale=1., size=100)
array_2 = np.random.normal(loc=0, scale=0.8,size=100)
print(np.var(array_1), np.var(array_2))
pg.homoscedasticity(array_1, array_2, alpha=.05)
1.2729265592243306 0.6022425373276372
(False, 0.0)
Parametric tests¶
Student’s t-test: the simplest statistical test¶
1-sample t-test: testing the value of a population mean¶
tests if the population mean of data is likely to be equal to a given value (technically if observations are drawn from a Gaussian distributions of given population mean).
pingouin.ttest returns the T_statistic, the p-value, the [degrees of freedom](https://en.wikipedia.org/wiki/Degrees_of_freedom_(statistics), the Cohen d effect size, the achieved [power](https://en.wikipedia.org/wiki/Power_(statistics)) of the test ( = 1 - type II error (beta) = P(Reject H0|H1 is true)), and the Bayes Factor of the alternative hypothesis
pg.ttest(data['MFQ'],0)
| T | p-val | dof | tail | cohen-d | power | BF10 | |
|---|---|---|---|---|---|---|---|
| T-test | 7.063 | 8.228161e-09 | 45 | two-sided | 1.041 | 1.0 | 1527145.713 |
2-sample t-test: testing for difference across populations¶
We have seen above that the mean MFQ in the MDD and HV populations
were different. To test if this is significant, we do a 2-sample t-test:
MDD_MFQ = data[data['group'] == 'MDD']['MFQ']
HV_MFQ = data[data['group'] == 'HV']['MFQ']
pg.ttest(MDD_MFQ, HV_MFQ)
| T | p-val | dof | dof-corr | tail | cohen-d | power | BF10 | |
|---|---|---|---|---|---|---|---|---|
| T-test | 5.866 | 6.718419e-07 | 44 | 40.991061 | two-sided | 1.486 | 0.997 | 22256.489 |
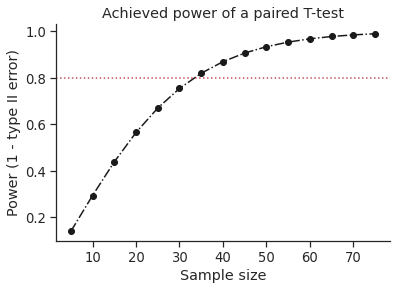
Plot achieved power of a paired T-test¶
Plot the curve of achieved power given the effect size (Cohen d) and the sample size of a paired T-test.
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style='ticks', context='notebook', font_scale=1.2)
d = 0.5 # Fixed effect size
n = np.arange(5, 80, 5) # Incrementing sample size
# Compute the achieved power
pwr = pg.power_ttest(d=d, n=n, contrast='paired', tail='two-sided')
# Start the plot
plt.plot(n, pwr, 'ko-.')
plt.axhline(0.8, color='r', ls=':')
plt.xlabel('Sample size')
plt.ylabel('Power (1 - type II error)')
plt.title('Achieved power of a paired T-test')
sns.despine()
Non parametric tests:¶
Unlike the parametric test these do not require the assumption of normal distributions.
“Mann-Whitney U Test (= Wilcoxon rank-sum test). It is the non-parametric version of the independent T-test.
Mwu tests the hypothesis that data in x and y are samples from continuous distributions with equal medians. The test assumes that x and y are independent. This test corrects for ties and by default uses a continuity correction.” - mwu-function
Test summary
‘W-val’ : W-value
‘p-val’ : p-value
‘RBC’ : matched pairs rank-biserial correlation (effect size)
‘CLES’ : common language effect size
pg.mwu(MDD_MFQ, HV_MFQ)
| U-val | p-val | RBC | CLES | |
|---|---|---|---|---|
| MWU | 442.0 | 0.000008 | -0.793103 | 0.872211 |
“Wilcoxon signed-rank test is the non-parametric version of the paired T-test.
The Wilcoxon signed-rank test tests the null hypothesis that two related paired samples come from the same distribution. A continuity correction is applied by default.” - wilcoxon - func
# example from the function definition
# Wilcoxon test on two related samples.
x = [20, 22, 19, 20, 22, 18, 24, 20]
y = [38, 37, 33, 29, 14, 12, 20, 22]
print("Medians = %.2f - %.2f" % (np.median(x), np.median(y)))
pg.wilcoxon(x, y, tail='two-sided')
Medians = 20.00 - 25.50
/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/scipy/stats/morestats.py:2879: UserWarning: Sample size too small for normal approximation.
warnings.warn("Sample size too small for normal approximation.")
| W-val | p-val | RBC | CLES | |
|---|---|---|---|---|
| Wilcoxon | 9.0 | 0.043089 | 0.5 | 0.609375 |

scipy.stats - Hypothesis testing: comparing two groups¶
For simple statistical tests, it is also possible to use the scipy.stats sub-module of scipy.
from scipy import stats
1-sample t-test: testing the value of a population mean¶
scipy.stats.ttest_1samp tests if the population mean of data is likely to be equal to a given value (technically if observations are drawn from a Gaussian distributions of given population mean). It returns the T statistic, and the p-value (see the function’s help):
stats.ttest_1samp(data['MFQ'].dropna(), 0)
Ttest_1sampResult(statistic=7.062650755155121, pvalue=8.228161269771835e-09)
With a p-value of 8^-09 we can claim that the population mean for the MFQ is not 0.
2-sample t-test: testing for difference across populations¶
We have seen above that the mean MFQ in the MDD and HV populations
were different. To test if this is significant, we do a 2-sample t-test
with scipy.stats.ttest_ind:
stats.ttest_ind(MDD_MFQ.dropna(), HV_MFQ.dropna())
Ttest_indResult(statistic=4.863337342528167, pvalue=1.5121907363581306e-05)
Paired tests: repeated measurements on the same indivuals¶
MFQ and SHAPS are measures of interest. Let us test if they are significantly different. We can use a 2 sample test:
stats.ttest_ind(data['MFQ'].dropna(), data['SHAPS'].dropna())
Ttest_indResult(statistic=-2.4632108396344363, pvalue=0.01567217083007739)
The problem with this approach is that it forgets that there are links
between observations: MFQ and SHAPS are measured on the same individuals.
Thus the variance due to inter-subject variability is confounding, and
can be removed, using a “paired test”, or “repeated measures test”:
stats.ttest_rel(data['MFQ'].dropna(), data['SHAPS'].dropna())
Ttest_relResult(statistic=-3.006858621511653, pvalue=0.004308118483154467)
This is equivalent to a 1-sample test on the difference::
stats.ttest_1samp(data['MFQ'].dropna() - data['SHAPS'].dropna(), 0)
Ttest_1sampResult(statistic=nan, pvalue=nan)
T-tests assume Gaussian errors. We can use a Wilcoxon signed-rank test, that relaxes this assumption:
stats.wilcoxon(data['MFQ'].dropna(), data['SHAPS'].dropna())
WilcoxonResult(statistic=177.0, pvalue=0.004985029827378135)
Note: The corresponding test in the non paired case is the Mann–Whitney U test, scipy.stats.mannwhitneyu.

statsmodels - use “formulas” to specify statistical models in Python¶
Use statsmodels to perform linear models, multiple factors or analysis of variance.
A simple linear regression¶
Given two set of observations, x and y, we want to test the hypothesis that y is a linear function of x.
In other terms:
\(y = x * coef + intercept + e\)
where \(e\) is observation noise. We will use the statsmodels module to:
Fit a linear model. We will use the simplest strategy, ordinary least squares (OLS).
Test that \(coef\) is non zero.
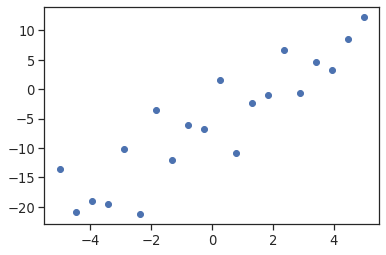
First, we generate simulated data according to the model:
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(-5, 5, 20)
np.random.seed(1)
# normal distributed noise
y = -5 + 3*x + 4 * np.random.normal(size=x.shape)
# Create a data frame containing all the relevant variables
data = pd.DataFrame({'x': x, 'y': y})
plt.plot(x, y, 'o');
Then we specify an OLS model and fit it:
from statsmodels.formula.api import ols
model = ols("y ~ x", data).fit()
Note: For more about “formulas” for statistics in Python, see the statsmodels documentation.
We can inspect the various statistics derived from the fit::
print(model.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.804
Model: OLS Adj. R-squared: 0.794
Method: Least Squares F-statistic: 74.03
Date: Sun, 17 Oct 2021 Prob (F-statistic): 8.56e-08
Time: 16:14:47 Log-Likelihood: -57.988
No. Observations: 20 AIC: 120.0
Df Residuals: 18 BIC: 122.0
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept -5.5335 1.036 -5.342 0.000 -7.710 -3.357
x 2.9369 0.341 8.604 0.000 2.220 3.654
==============================================================================
Omnibus: 0.100 Durbin-Watson: 2.956
Prob(Omnibus): 0.951 Jarque-Bera (JB): 0.322
Skew: -0.058 Prob(JB): 0.851
Kurtosis: 2.390 Cond. No. 3.03
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Terminology¶
Statsmodels uses a statistical terminology: the y variable in statsmodels is called endogenous while the x variable is called exogenous. This is discussed in more detail here. To simplify, y (endogenous) is the value you are trying to predict, while x (exogenous) represents the features you are using to make the prediction.
Categorical variables: comparing groups or multiple categories¶
Let us go back to our data. We can write a comparison between MFQ of MDD and HV groups using a linear model:
data = pd.read_csv('/data/participants_stats.tsv', sep='\t', na_values=".")
model = ols("MFQ ~ group + 1", data).fit()
print(model.summary())
OLS Regression Results
==============================================================================
Dep. Variable: MFQ R-squared: 0.350
Model: OLS Adj. R-squared: 0.335
Method: Least Squares F-statistic: 23.65
Date: Sun, 17 Oct 2021 Prob (F-statistic): 1.51e-05
Time: 16:14:54 Log-Likelihood: -133.38
No. Observations: 46 AIC: 270.8
Df Residuals: 44 BIC: 274.4
Df Model: 1
Covariance Type: nonrobust
================================================================================
coef std err t P>|t| [0.025 0.975]
--------------------------------------------------------------------------------
Intercept 1.5294 1.090 1.403 0.168 -0.668 3.727
group[T.MDD] 6.6775 1.373 4.863 0.000 3.910 9.445
==============================================================================
Omnibus: 2.463 Durbin-Watson: 1.836
Prob(Omnibus): 0.292 Jarque-Bera (JB): 1.930
Skew: 0.502 Prob(JB): 0.381
Kurtosis: 3.012 Cond. No. 3.05
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Tips on specifying model¶
Forcing categorical - the ‘group’ variable is automatically detected as a categorical variable, and thus each of its different values is treated as different entities.
An integer column can be forced to be treated as categorical using:
model = ols('MFQ ~ C(group)', data).fit()
Intercept: We can remove the intercept using - 1 in the formula, or force the use of an intercept using + 1.
Link to t-tests between different MFQ and SHAPS¶
To compare different types of our measurements of interest, we need to create a "long-form" table, listing them, where their type is indicated by a categorical variable:
data_mfq = pd.DataFrame({'MOI': data['MFQ'], 'type': 'MFQ'})
data_shaps = pd.DataFrame({'MOI': data['SHAPS'], 'type': 'SHAPS'})
data_long = pd.concat((data_mfq, data_shaps))
print(data_long[::8])
MOI type
0 0.0 MFQ
8 1.0 MFQ
16 4.0 MFQ
24 NaN MFQ
32 0.0 MFQ
40 0.0 MFQ
48 8.0 MFQ
5 14.0 SHAPS
13 2.0 SHAPS
21 9.0 SHAPS
29 8.0 SHAPS
37 14.0 SHAPS
45 1.0 SHAPS
model = ols("MOI ~ type", data_long).fit()
print(model.summary())
OLS Regression Results
==============================================================================
Dep. Variable: MOI R-squared: 0.063
Model: OLS Adj. R-squared: 0.053
Method: Least Squares F-statistic: 6.067
Date: Sun, 17 Oct 2021 Prob (F-statistic): 0.0157
Time: 16:14:56 Log-Likelihood: -304.93
No. Observations: 92 AIC: 613.9
Df Residuals: 90 BIC: 618.9
Df Model: 1
Covariance Type: nonrobust
=================================================================================
coef std err t P>|t| [0.025 0.975]
---------------------------------------------------------------------------------
Intercept 5.7391 0.992 5.784 0.000 3.768 7.710
type[T.SHAPS] 3.4565 1.403 2.463 0.016 0.669 6.244
==============================================================================
Omnibus: 6.740 Durbin-Watson: 1.892
Prob(Omnibus): 0.034 Jarque-Bera (JB): 6.017
Skew: 0.548 Prob(JB): 0.0494
Kurtosis: 2.391 Cond. No. 2.62
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
We can see that we retrieve the same values for t-test and corresponding p-values for the effect of the type of MOI than the previous t-test:
stats.ttest_ind(data['MFQ'].dropna(), data['SHAPS'].dropna())
Ttest_indResult(statistic=-2.4632108396344363, pvalue=0.01567217083007739)
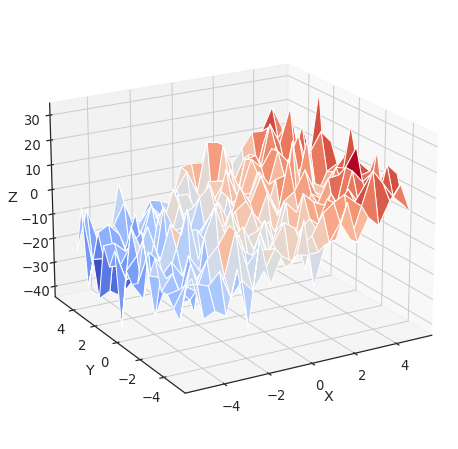
Multiple Regression: including multiple factors¶
Consider a linear model explaining a variable z (the dependent
variable) with 2 variables x and y:
\(z = x \, c_1 + y \, c_2 + i + e\)
Such a model can be seen in 3D as fitting a plane to a cloud of (x,
y, z) points (see the following figure).
from mpl_toolkits.mplot3d import Axes3D
x = np.linspace(-5, 5, 21)
# We generate a 2D grid
X, Y = np.meshgrid(x, x)
# To get reproducable values, provide a seed value
np.random.seed(1)
# Z is the elevation of this 2D grid
Z = -5 + 3*X - 0.5*Y + 8 * np.random.normal(size=X.shape)
# Plot the data
fig = plt.figure(figsize=(8, 8))
ax = fig.gca(projection='3d')
surf = ax.plot_surface(X, Y, Z, cmap=plt.cm.coolwarm,
rstride=1, cstride=1)
ax.view_init(20, -120)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
Text(0.5, 0, 'Z')
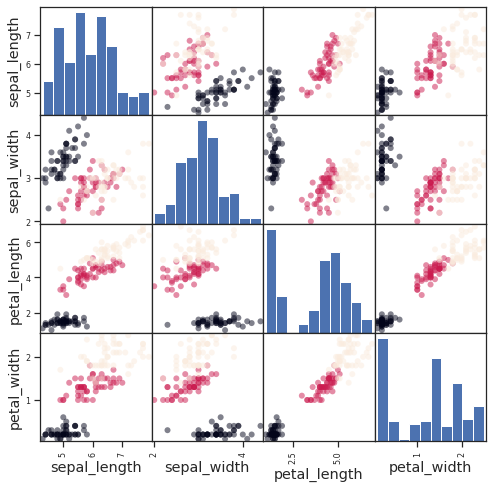
Example: the iris dataset¶
Sepal and petal size tend to be related: bigger flowers are bigger! But is there, in addition, a systematic effect of species?
from pandas.plotting import scatter_matrix
#Load the data
data = sns.load_dataset('iris')
# Express the names as categories
categories = pd.Categorical(data['species'])
# The parameter 'c' is passed to plt.scatter and will control the color
scatter_matrix(data, c=categories.codes, marker='o', figsize=(8, 8))
# Plot figure
fig.suptitle("blue: setosa, green: versicolor, red: virginica", size=13)
plt.show()
model = ols('sepal_width ~ species + petal_length', data).fit()
print(model.summary())
OLS Regression Results
==============================================================================
Dep. Variable: sepal_width R-squared: 0.486
Model: OLS Adj. R-squared: 0.476
Method: Least Squares F-statistic: 46.08
Date: Sun, 17 Oct 2021 Prob (F-statistic): 5.14e-21
Time: 16:14:59 Log-Likelihood: -37.808
No. Observations: 150 AIC: 83.62
Df Residuals: 146 BIC: 95.66
Df Model: 3
Covariance Type: nonrobust
=========================================================================================
coef std err t P>|t| [0.025 0.975]
-----------------------------------------------------------------------------------------
Intercept 2.9919 0.099 30.206 0.000 2.796 3.188
species[T.versicolor] -1.4927 0.181 -8.264 0.000 -1.850 -1.136
species[T.virginica] -1.6741 0.255 -6.557 0.000 -2.179 -1.170
petal_length 0.2983 0.060 4.932 0.000 0.179 0.418
==============================================================================
Omnibus: 3.167 Durbin-Watson: 1.771
Prob(Omnibus): 0.205 Jarque-Bera (JB): 3.214
Skew: -0.121 Prob(JB): 0.200
Kurtosis: 3.675 Cond. No. 54.0
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Post-hoc hypothesis testing: analysis of variance (ANOVA)¶
In the above iris example, we wish to test if the petal length is different between versicolor and virginica, after removing the effect of sepal width. This can be formulated as testing the difference between the coefficient associated to versicolor and virginica in the linear model estimated above (it is an Analysis of Variance, ANOVA. For this, we write a vector of ‘contrast’ on the parameters estimated: we want to test "name[T.versicolor] - name[T.virginica]", with an F-test:
print(model.f_test([0, 1, -1, 0]))
<F test: F=array([[3.26191465]]), p=0.07296614041660528, df_denom=146, df_num=1>
Is this difference significant?

seaborn - use visualization for statistical exploration¶
Seaborn combines simple statistical fits with plotting on pandas dataframes.
Let us consider a data giving wages and many other personal information on 500 individuals (Berndt, ER. The Practice of Econometrics. 1991. NY:Addison-Wesley) which we will gather using a small function based on the scipy tutorials.
def download_wages_data():
import urllib
import os
import pandas as pd
if not os.path.exists('wages.txt'):
# Download the file if it is not present
urllib.request.urlretrieve('http://lib.stat.cmu.edu/datasets/CPS_85_Wages',
'wages.txt')
# Give names to the columns
names = [
'EDUCATION: Number of years of education',
'SOUTH: 1=Person lives in South, 0=Person lives elsewhere',
'SEX: 1=Female, 0=Male',
'EXPERIENCE: Number of years of work experience',
'UNION: 1=Union member, 0=Not union member',
'WAGE: Wage (dollars per hour)',
'AGE: years',
'RACE: 1=Other, 2=Hispanic, 3=White',
'OCCUPATION: 1=Management, 2=Sales, 3=Clerical, 4=Service, 5=Professional, 6=Other',
'SECTOR: 0=Other, 1=Manufacturing, 2=Construction',
'MARR: 0=Unmarried, 1=Married',
]
short_names = [n.split(':')[0] for n in names]
data = pd.read_csv('wages.txt', skiprows=27, skipfooter=6, sep=None,
header=None)
data.columns = short_names
# Log-transform the wages, because they typically are increased with
# multiplicative factors
import numpy as np
data['WAGE'] = np.log10(data['WAGE'])
return data
import pandas as pd
data = download_wages_data()
data.head()
/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/ipykernel_launcher.py:31: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support skipfooter; you can avoid this warning by specifying engine='python'.
| EDUCATION | SOUTH | SEX | EXPERIENCE | UNION | WAGE | AGE | RACE | OCCUPATION | SECTOR | MARR | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 8 | 0 | 1 | 21 | 0 | 0.707570 | 35 | 2 | 6 | 1 | 1 |
| 1 | 9 | 0 | 1 | 42 | 0 | 0.694605 | 57 | 3 | 6 | 1 | 1 |
| 2 | 12 | 0 | 0 | 1 | 0 | 0.824126 | 19 | 3 | 6 | 1 | 0 |
| 3 | 12 | 0 | 0 | 4 | 0 | 0.602060 | 22 | 3 | 6 | 0 | 0 |
| 4 | 12 | 0 | 0 | 17 | 0 | 0.875061 | 35 | 3 | 6 | 0 | 1 |
Pairplot: scatter matrices¶
We can easily have an intuition on the interactions between continuous variables using seaborn.pairplot to display a scatter matrix:
import seaborn
seaborn.set()
seaborn.pairplot(data, vars=['WAGE', 'AGE', 'EDUCATION'], kind='reg')
<seaborn.axisgrid.PairGrid at 0x7fe5c444a350>
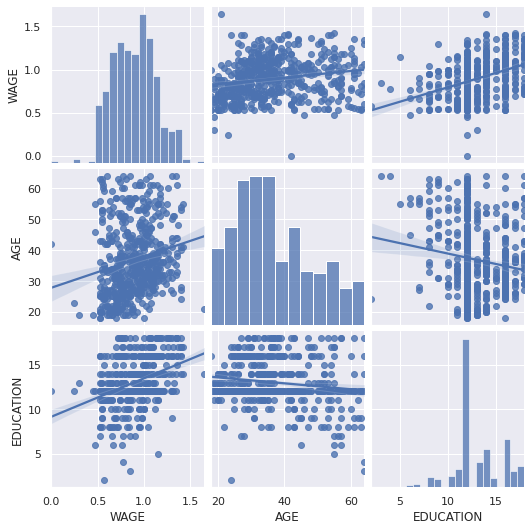
Categorical variables can be plotted as the hue:
seaborn.pairplot(data, vars=['WAGE', 'AGE', 'EDUCATION'], kind='reg', hue='SEX')
<seaborn.axisgrid.PairGrid at 0x7fe5c2183e10>
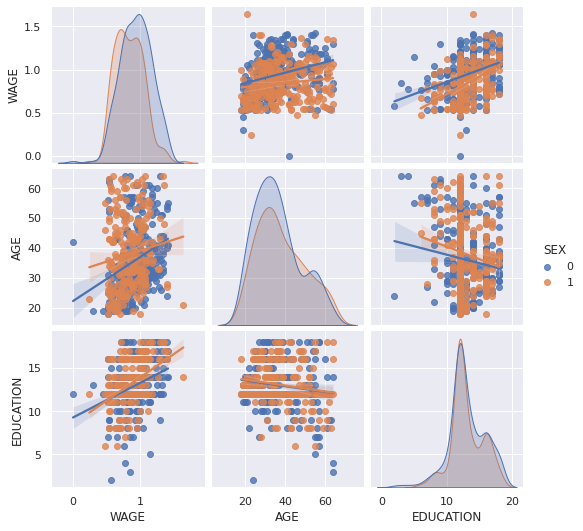
lmplot: plotting a univariate regression¶
A regression capturing the relation between one variable and another, e.g. wage and eduction, can be plotted using seaborn.lmplot:
seaborn.lmplot(y='WAGE', x='EDUCATION', data=data)
<seaborn.axisgrid.FacetGrid at 0x7fe5c23700d0>
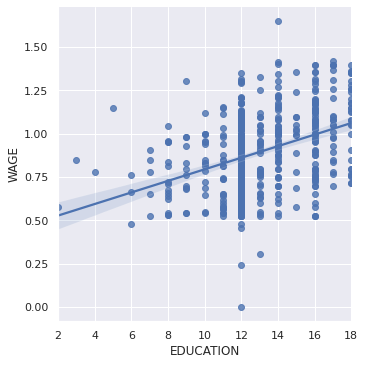
Robust regression¶
Given that, in the above plot, there seems to be a couple of data points that are outside of the main cloud to the right, they might be outliers, not representative of the population, but driving the regression.
To compute a regression that is less sensitive to outliers, one must use a robust model. This is done in seaborn using robust=True in the plotting functions, or in statsmodels by replacing the use of the OLS by a “Robust Linear Model”, statsmodels.formula.api.rlm.
Testing for interactions¶
Do wages increase more with education for people with dark hair than with light hair?
seaborn.lmplot(y='WAGE', x='EDUCATION', hue='SEX', data=data)
<seaborn.axisgrid.FacetGrid at 0x7fe5caa1f9d0>
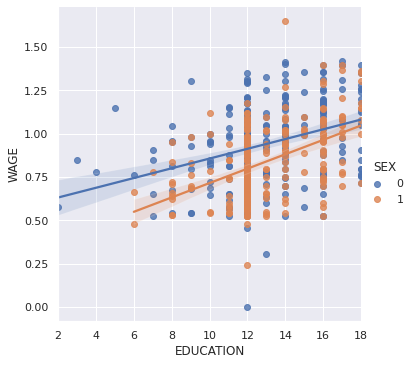
The plot above is made of two different fits. We need to formulate a single model that tests for a variance of slope across the population. This is done via an “interaction”.
from statsmodels.formula.api import ols
result = ols(formula='WAGE ~ EDUCATION + SEX + EDUCATION * SEX', data=data).fit()
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: WAGE R-squared: 0.198
Model: OLS Adj. R-squared: 0.194
Method: Least Squares F-statistic: 43.72
Date: Sun, 17 Oct 2021 Prob (F-statistic): 2.94e-25
Time: 16:15:09 Log-Likelihood: 88.503
No. Observations: 534 AIC: -169.0
Df Residuals: 530 BIC: -151.9
Df Model: 3
Covariance Type: nonrobust
=================================================================================
coef std err t P>|t| [0.025 0.975]
---------------------------------------------------------------------------------
Intercept 0.5748 0.058 9.861 0.000 0.460 0.689
EDUCATION 0.0281 0.004 6.411 0.000 0.019 0.037
SEX -0.2750 0.093 -2.972 0.003 -0.457 -0.093
EDUCATION:SEX 0.0134 0.007 1.919 0.056 -0.000 0.027
==============================================================================
Omnibus: 4.838 Durbin-Watson: 1.825
Prob(Omnibus): 0.089 Jarque-Bera (JB): 5.000
Skew: -0.156 Prob(JB): 0.0821
Kurtosis: 3.356 Cond. No. 170.
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Can we conclude that education benefits people with dark hair more than people with light hair?
Take home messages¶
Hypothesis testing and p-value give you the significance of an effect / difference
Formulas (with categorical variables) enable you to express rich links in your data
Visualizing your data and simple model fits matters!
Conditioning (adding factors that can explain all or part of the variation) is an important modeling aspect that changes the interpretation.