Nipype Quickstart - non-imaging¶
This is a very quick non-imaging introduction to Nipype workflows. For a more comprehensive introduction, check the next section of the tutorial.
This notebook is taken from reproducible-imaging repository
Import a few things from nipype¶
import os
from nipype import Workflow, Node, Function
Creating Workflow with one Node that adds two numbers
def sum(a, b):
return a + b
wf = Workflow('hello')
adder = Node(Function(input_names=['a', 'b'],
output_names=['sum'],
function=sum),
name='a_plus_b')
adder.inputs.a = 1
adder.inputs.b = 3
wf.add_nodes([adder])
wf.base_dir = os.getcwd()
eg = wf.run()
list(eg.nodes())[0].result.outputs
211017-18:07:54,305 nipype.workflow INFO:
Workflow hello settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:54,339 nipype.workflow INFO:
Running serially.
211017-18:07:54,340 nipype.workflow INFO:
[Node] Setting-up "hello.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/a_plus_b".
211017-18:07:54,377 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:54,434 nipype.workflow INFO:
[Node] Finished "hello.a_plus_b".
sum = 4
Creating a second node and connecting to the hello Workflow
def concat(a, b):
return [a, b]
concater = Node(Function(input_names=['a', 'b'],
output_names=['some_list'],
function=concat),
name='concat_a_b')
wf.connect(adder, 'sum', concater, 'a')
concater.inputs.b = 3
eg = wf.run()
print(eg.nodes())
211017-18:07:54,468 nipype.workflow INFO:
Workflow hello settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:54,516 nipype.workflow INFO:
Running serially.
211017-18:07:54,518 nipype.workflow INFO:
[Node] Setting-up "hello.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/a_plus_b".
211017-18:07:54,526 nipype.workflow INFO:
[Node] Outdated cache found for "hello.a_plus_b".
211017-18:07:54,565 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:54,603 nipype.workflow INFO:
[Node] Finished "hello.a_plus_b".
211017-18:07:54,605 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/concat_a_b".
211017-18:07:54,640 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:54,698 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
[hello.a_plus_b, hello.concat_a_b]
And we can check results of our Workflow, we should see a list:
list(eg.nodes())[-1].result.outputs
some_list = [4, 3]
We will try to add additional Node that adds one:
def plus_one(a):
return a + 1
plusone = Node(Function(input_names=['a'],
output_names=['out'],
function=plus_one),
name='add_1')
wf.connect(concater, 'some_list', plusone, 'a')
try:
eg = wf.run()
except(RuntimeError) as err:
print("RuntimeError:", err)
else:
raise
211017-18:07:54,731 nipype.workflow INFO:
Workflow hello settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:54,868 nipype.workflow INFO:
Running serially.
211017-18:07:54,869 nipype.workflow INFO:
[Node] Setting-up "hello.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/a_plus_b".
211017-18:07:54,907 nipype.workflow INFO:
[Node] Cached "hello.a_plus_b" - collecting precomputed outputs
211017-18:07:54,908 nipype.workflow INFO:
[Node] "hello.a_plus_b" found cached.
211017-18:07:54,910 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/concat_a_b".
211017-18:07:54,922 nipype.workflow INFO:
[Node] Outdated cache found for "hello.concat_a_b".
211017-18:07:54,958 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:55,13 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:55,14 nipype.workflow INFO:
[Node] Setting-up "hello.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/add_1".
211017-18:07:55,50 nipype.workflow INFO:
[Node] Running "add_1" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:55,55 nipype.workflow WARNING:
Storing result file without outputs
211017-18:07:55,60 nipype.workflow WARNING:
[Node] Error on "hello.add_1" (/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/add_1)
211017-18:07:55,74 nipype.workflow ERROR:
Node add_1 failed to run on host a54b8d983419.
211017-18:07:55,78 nipype.workflow ERROR:
Saving crash info to /home/neuro/workshop_weizmann/workshop/nipype/notebooks/crash-20211017-180755-neuro-add_1-7b3ed9c1-2c8c-44b6-9693-65e906a76506.pklz
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/pipeline/plugins/linear.py", line 46, in run
node.run(updatehash=updatehash)
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 516, in run
result = self._run_interface(execute=True)
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 635, in _run_interface
return self._run_command(execute)
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 741, in _run_command
result = self._interface.run(cwd=outdir)
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/interfaces/base/core.py", line 419, in run
runtime = self._run_interface(runtime)
File "/opt/miniconda-latest/envs/neuro/lib/python3.7/site-packages/nipype/interfaces/utility/wrappers.py", line 142, in _run_interface
out = function_handle(**args)
File "<string>", line 2, in plus_one
TypeError: can only concatenate list (not "int") to list
211017-18:07:55,86 nipype.workflow INFO:
***********************************
211017-18:07:55,87 nipype.workflow ERROR:
could not run node: hello.add_1
211017-18:07:55,88 nipype.workflow INFO:
crashfile: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/crash-20211017-180755-neuro-add_1-7b3ed9c1-2c8c-44b6-9693-65e906a76506.pklz
211017-18:07:55,91 nipype.workflow INFO:
***********************************
RuntimeError: Workflow did not execute cleanly. Check log for details
This time the workflow didn’t execute cleanly and we got an error. We can use nipypecli to read the crashfile (note, that if you have multiple crashfiles in the directory you’ll have to provide a full name):
!nipypecli crash crash*
Usage: nipypecli crash [OPTIONS] CRASHFILE
Try 'nipypecli crash -h' for help.
Error: Got unexpected extra arguments (crash-20211017-171039-neuro-selectfiles-f56abef1-3008-4e84-941c-c2800895302b.txt crash-20211017-180755-neuro-add_1-7b3ed9c1-2c8c-44b6-9693-65e906a76506.pklz)
It clearly shows the problematic Node and its input. We tried to add an integer to a list, this operation is not allowed in Python.
Let’s try using MapNode
from nipype import MapNode
plusone = MapNode(Function(input_names=['a'],
output_names=['out'],
function=plus_one),
iterfield=['a'],
name='add_1')
wf = Workflow('hello_mapnode')
adder = Node(Function(input_names=['a', 'b'],
output_names=['sum'],
function=sum),
name='a_plus_b')
adder.inputs.a = 1
adder.inputs.b = 3
wf.connect(adder, 'sum', concater, 'a')
concater.inputs.b = 3
wf.connect(concater, 'some_list', plusone, 'a')
wf.base_dir = os.getcwd()
eg = wf.run()
print(eg.nodes())
211017-18:07:56,314 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:56,382 nipype.workflow INFO:
Running serially.
211017-18:07:56,383 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/a_plus_b".
211017-18:07:56,405 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,448 nipype.workflow INFO:
[Node] Finished "hello_mapnode.a_plus_b".
211017-18:07:56,449 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/concat_a_b".
211017-18:07:56,476 nipype.workflow INFO:
[Node] Cached "hello.concat_a_b" - collecting precomputed outputs
211017-18:07:56,477 nipype.workflow INFO:
[Node] "hello.concat_a_b" found cached.
211017-18:07:56,479 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/add_1".
211017-18:07:56,512 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/add_1/mapflow/_add_10".
211017-18:07:56,538 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,577 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:56,585 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/add_1/mapflow/_add_11".
211017-18:07:56,606 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,643 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:56,661 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
[hello_mapnode.a_plus_b, hello.concat_a_b, hello_mapnode.add_1]
Now the workflow finished without problems, let’s see the results from hello.add_1:
print(list(eg.nodes())[2].result.outputs)
Bunch(out=[5, 4])
And now we will run the example with iterables:
adder.iterables = ('a', [1, 2])
adder.inputs.b = 2
eg = wf.run()
print(eg.nodes())
211017-18:07:56,695 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:56,742 nipype.workflow INFO:
Running serially.
211017-18:07:56,743 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/a_plus_b".
211017-18:07:56,765 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,796 nipype.workflow INFO:
[Node] Finished "hello_mapnode.a_plus_b".
211017-18:07:56,797 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_2/concat_a_b".
211017-18:07:56,827 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,864 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:56,866 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/add_1".
211017-18:07:56,894 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/add_1/mapflow/_add_10".
211017-18:07:56,915 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:56,949 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:56,958 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/add_1/mapflow/_add_11".
211017-18:07:56,981 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,16 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:57,34 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:07:57,36 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/a_plus_b".
211017-18:07:57,53 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,91 nipype.workflow INFO:
[Node] Finished "hello_mapnode.a_plus_b".
211017-18:07:57,92 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_1/concat_a_b".
211017-18:07:57,117 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,148 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:57,150 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/add_1".
211017-18:07:57,181 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/add_1/mapflow/_add_10".
211017-18:07:57,197 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,231 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:57,238 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/add_1/mapflow/_add_11".
211017-18:07:57,256 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,296 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:57,309 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
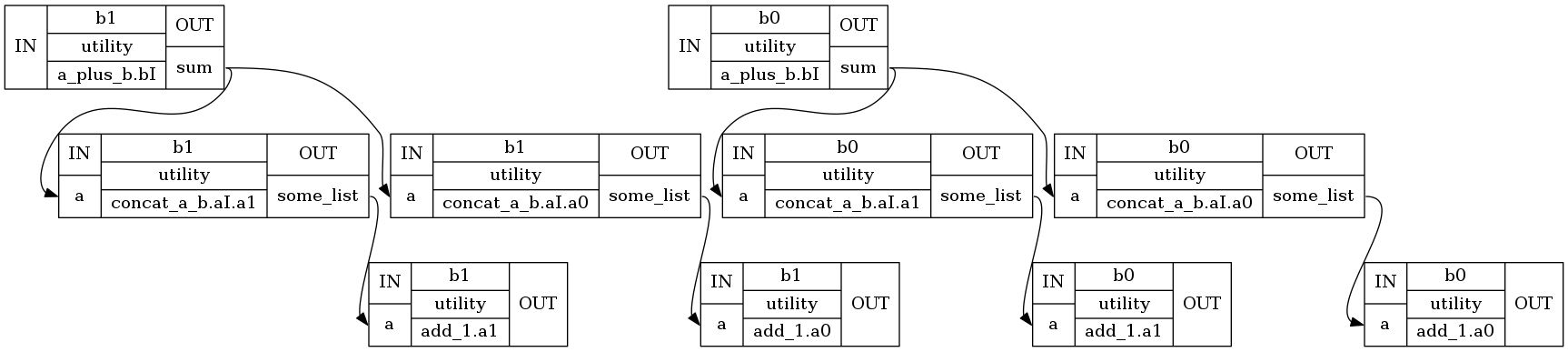
[hello_mapnode.a_plus_b.aI.a0, hello.concat_a_b.a0, hello_mapnode.add_1.a0, hello_mapnode.a_plus_b.aI.a1, hello.concat_a_b.a1, hello_mapnode.add_1.a1]
Now we have 6 nodes, we can check results for hello.add_1.a1
list(eg.nodes())[5].result.outputs
Bunch(out=[5, 4])
wf.write_graph(graph2use='exec')
211017-18:07:57,609 nipype.workflow INFO:
Generated workflow graph: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png (graph2use=exec, simple_form=True).
'/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png'
from IPython.display import Image
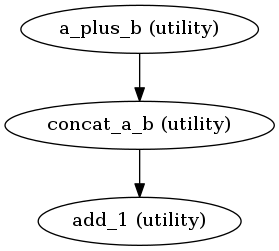
We can plot a general structure of the workflow:
Image("hello_mapnode/graph.png")
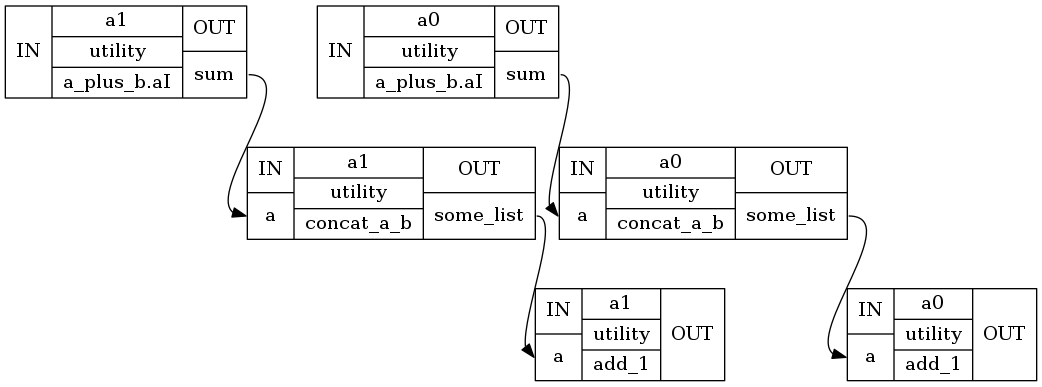
And more detailed structure with all nodes:
Image("hello_mapnode/graph_detailed.png")
We will introduce another iterables, for the concater Node:
concater.iterables = ('b', [3, 4])
eg = wf.run()
eg.nodes();
211017-18:07:57,705 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:57,801 nipype.workflow INFO:
Running serially.
211017-18:07:57,802 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/a_plus_b".
211017-18:07:57,828 nipype.workflow INFO:
[Node] Cached "hello_mapnode.a_plus_b" - collecting precomputed outputs
211017-18:07:57,829 nipype.workflow INFO:
[Node] "hello_mapnode.a_plus_b" found cached.
211017-18:07:57,831 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_2/_b_4/concat_a_b".
211017-18:07:57,870 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:57,914 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:57,915 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1".
211017-18:07:57,962 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1/mapflow/_add_10".
211017-18:07:57,989 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,39 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:58,49 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1/mapflow/_add_11".
211017-18:07:58,76 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,129 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:58,154 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:07:58,156 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_2/_b_3/concat_a_b".
211017-18:07:58,199 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,246 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:58,247 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1".
211017-18:07:58,297 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1/mapflow/_add_10".
211017-18:07:58,333 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,373 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:58,390 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1/mapflow/_add_11".
211017-18:07:58,521 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,592 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:58,617 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:07:58,619 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/a_plus_b".
211017-18:07:58,642 nipype.workflow INFO:
[Node] Cached "hello_mapnode.a_plus_b" - collecting precomputed outputs
211017-18:07:58,643 nipype.workflow INFO:
[Node] "hello_mapnode.a_plus_b" found cached.
211017-18:07:58,644 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_1/_b_4/concat_a_b".
211017-18:07:58,687 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,739 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:58,740 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1".
211017-18:07:58,789 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1/mapflow/_add_10".
211017-18:07:58,823 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,869 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:58,881 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1/mapflow/_add_11".
211017-18:07:58,925 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:58,988 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:59,17 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:07:59,18 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_1/_b_3/concat_a_b".
211017-18:07:59,58 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:59,109 nipype.workflow INFO:
[Node] Finished "hello.concat_a_b".
211017-18:07:59,110 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1".
211017-18:07:59,160 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1/mapflow/_add_10".
211017-18:07:59,197 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:59,258 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:07:59,272 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1/mapflow/_add_11".
211017-18:07:59,305 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:07:59,360 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:07:59,385 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
wf.write_graph(graph2use='exec')
211017-18:07:59,688 nipype.workflow INFO:
Generated workflow graph: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png (graph2use=exec, simple_form=True).
'/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png'
Image("hello_mapnode/graph_detailed.png")
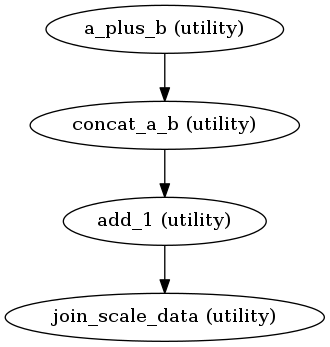
Now we will introduce JoinNode that allows us to merge results together:
def merge_and_scale_data(data2):
import numpy as np
return (np.array(data2) * 1000).tolist()
from nipype import JoinNode
joiner = JoinNode(Function(input_names=['data2'],
output_names=['data_scaled'],
function=merge_and_scale_data),
name='join_scale_data',
joinsource=adder,
joinfield=['data2'])
wf.connect(plusone, 'out', joiner, 'data2')
eg = wf.run()
eg.nodes()
211017-18:07:59,729 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:07:59,859 nipype.workflow INFO:
Running serially.
211017-18:07:59,861 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/a_plus_b".
211017-18:07:59,884 nipype.workflow INFO:
[Node] Cached "hello_mapnode.a_plus_b" - collecting precomputed outputs
211017-18:07:59,886 nipype.workflow INFO:
[Node] "hello_mapnode.a_plus_b" found cached.
211017-18:07:59,887 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_2/_b_4/concat_a_b".
211017-18:07:59,925 nipype.workflow INFO:
[Node] Cached "hello.concat_a_b" - collecting precomputed outputs
211017-18:07:59,926 nipype.workflow INFO:
[Node] "hello.concat_a_b" found cached.
211017-18:07:59,929 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1".
211017-18:07:59,963 nipype.workflow INFO:
[Node] Outdated cache found for "hello_mapnode.add_1".
211017-18:08:00,41 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1/mapflow/_add_10".
211017-18:08:00,70 nipype.workflow INFO:
[Node] Outdated cache found for "_add_10".
211017-18:08:00,127 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:00,179 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:08:00,191 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_4/add_1/mapflow/_add_11".
211017-18:08:00,202 nipype.workflow INFO:
[Node] Outdated cache found for "_add_11".
211017-18:08:00,248 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:00,299 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:08:00,321 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:08:00,324 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_2/_b_3/concat_a_b".
211017-18:08:00,365 nipype.workflow INFO:
[Node] Cached "hello.concat_a_b" - collecting precomputed outputs
211017-18:08:00,366 nipype.workflow INFO:
[Node] "hello.concat_a_b" found cached.
211017-18:08:00,368 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1".
211017-18:08:00,410 nipype.workflow INFO:
[Node] Outdated cache found for "hello_mapnode.add_1".
211017-18:08:00,464 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1/mapflow/_add_10".
211017-18:08:00,473 nipype.workflow INFO:
[Node] Outdated cache found for "_add_10".
211017-18:08:00,538 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:00,609 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:08:00,634 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_2/_b_3/add_1/mapflow/_add_11".
211017-18:08:00,655 nipype.workflow INFO:
[Node] Outdated cache found for "_add_11".
211017-18:08:00,783 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:00,838 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:08:00,866 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:08:00,868 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/a_plus_b".
211017-18:08:00,903 nipype.workflow INFO:
[Node] Cached "hello_mapnode.a_plus_b" - collecting precomputed outputs
211017-18:08:00,904 nipype.workflow INFO:
[Node] "hello_mapnode.a_plus_b" found cached.
211017-18:08:00,905 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_1/_b_4/concat_a_b".
211017-18:08:00,963 nipype.workflow INFO:
[Node] Cached "hello.concat_a_b" - collecting precomputed outputs
211017-18:08:00,965 nipype.workflow INFO:
[Node] "hello.concat_a_b" found cached.
211017-18:08:00,966 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1".
211017-18:08:01,7 nipype.workflow INFO:
[Node] Outdated cache found for "hello_mapnode.add_1".
211017-18:08:01,88 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1/mapflow/_add_10".
211017-18:08:01,104 nipype.workflow INFO:
[Node] Outdated cache found for "_add_10".
211017-18:08:01,180 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:01,265 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:08:01,280 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_4/add_1/mapflow/_add_11".
211017-18:08:01,297 nipype.workflow INFO:
[Node] Outdated cache found for "_add_11".
211017-18:08:01,369 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:01,452 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:08:01,481 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:08:01,483 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.join_scale_data" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_b_4/join_scale_data".
211017-18:08:01,551 nipype.workflow INFO:
[Node] Running "join_scale_data" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:01,609 nipype.workflow INFO:
[Node] Finished "hello_mapnode.join_scale_data".
211017-18:08:01,610 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello/_a_1/_b_3/concat_a_b".
211017-18:08:01,658 nipype.workflow INFO:
[Node] Cached "hello.concat_a_b" - collecting precomputed outputs
211017-18:08:01,658 nipype.workflow INFO:
[Node] "hello.concat_a_b" found cached.
211017-18:08:01,660 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.add_1" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1".
211017-18:08:01,688 nipype.workflow INFO:
[Node] Outdated cache found for "hello_mapnode.add_1".
211017-18:08:01,750 nipype.workflow INFO:
[Node] Setting-up "_add_10" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1/mapflow/_add_10".
211017-18:08:01,759 nipype.workflow INFO:
[Node] Outdated cache found for "_add_10".
211017-18:08:01,833 nipype.workflow INFO:
[Node] Running "_add_10" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:01,900 nipype.workflow INFO:
[Node] Finished "_add_10".
211017-18:08:01,912 nipype.workflow INFO:
[Node] Setting-up "_add_11" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_a_1/_b_3/add_1/mapflow/_add_11".
211017-18:08:01,919 nipype.workflow INFO:
[Node] Outdated cache found for "_add_11".
211017-18:08:01,976 nipype.workflow INFO:
[Node] Running "_add_11" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:02,33 nipype.workflow INFO:
[Node] Finished "_add_11".
211017-18:08:02,59 nipype.workflow INFO:
[Node] Finished "hello_mapnode.add_1".
211017-18:08:02,61 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.join_scale_data" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/_b_3/join_scale_data".
211017-18:08:02,160 nipype.workflow INFO:
[Node] Running "join_scale_data" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:02,211 nipype.workflow INFO:
[Node] Finished "hello_mapnode.join_scale_data".
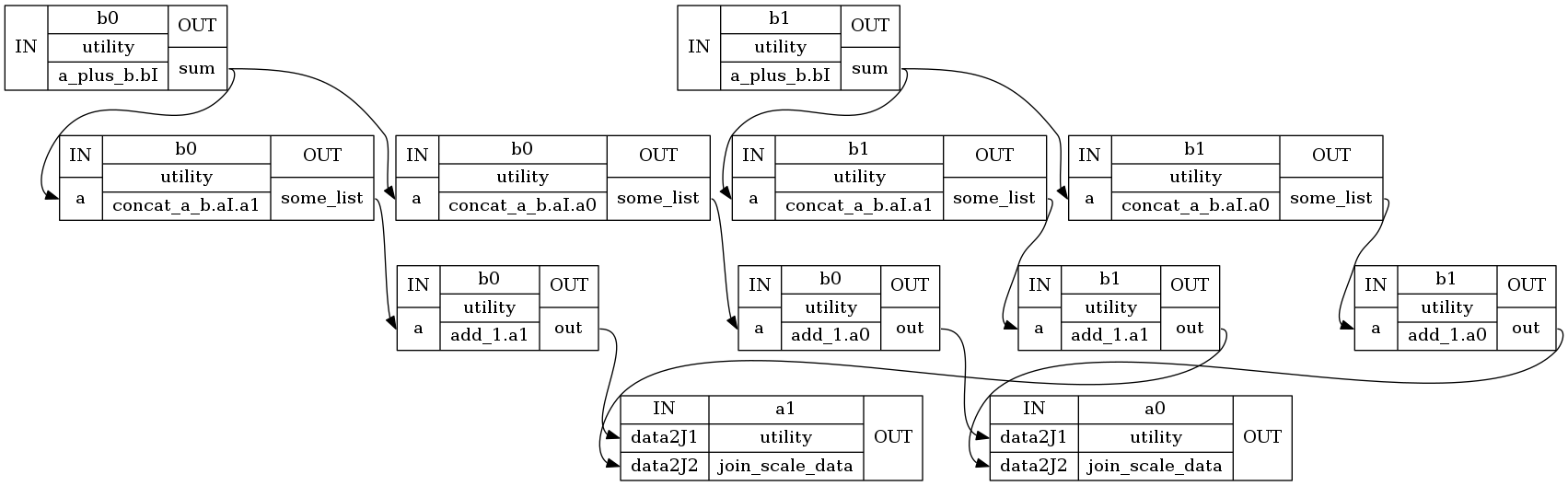
NodeView((hello_mapnode.join_scale_data.a0, hello_mapnode.join_scale_data.a1, hello_mapnode.a_plus_b.bI.b0, hello.concat_a_b.aI.a0.b0, hello_mapnode.add_1.a0.b0, hello.concat_a_b.aI.a1.b0, hello_mapnode.add_1.a1.b0, hello_mapnode.a_plus_b.bI.b1, hello.concat_a_b.aI.a0.b1, hello_mapnode.add_1.a0.b1, hello.concat_a_b.aI.a1.b1, hello_mapnode.add_1.a1.b1))
Let’s check the output of hello.join_scale_data.a0 node:
list(eg.nodes())[0].result.outputs
data_scaled = [[4000, 4000], [5000, 4000]]
wf.write_graph(graph2use='exec')
211017-18:08:02,553 nipype.workflow INFO:
Generated workflow graph: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png (graph2use=exec, simple_form=True).
'/home/neuro/workshop_weizmann/workshop/nipype/notebooks/hello_mapnode/graph.png'
Image("hello_mapnode/graph.png")
Image("hello_mapnode/graph_detailed.png")
%time eg = wf.run(plugin='MultiProc', plugin_args={'n_procs': 2})
211017-18:08:02,625 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:08:02,754 nipype.workflow INFO:
Running in parallel.
211017-18:08:02,761 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:02,811 nipype.workflow INFO:
[Job 0] Cached (hello_mapnode.a_plus_b).
211017-18:08:02,828 nipype.workflow INFO:
[Job 5] Cached (hello_mapnode.a_plus_b).
211017-18:08:04,763 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:04,810 nipype.workflow INFO:
[Job 1] Cached (hello.concat_a_b).
211017-18:08:04,834 nipype.workflow INFO:
[Job 3] Cached (hello.concat_a_b).
211017-18:08:06,856 nipype.workflow INFO:
[Job 6] Cached (hello.concat_a_b).
211017-18:08:06,890 nipype.workflow INFO:
[Job 9] Cached (hello.concat_a_b).
211017-18:08:08,773 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 6 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:08,887 nipype.workflow INFO:
[Job 12] Cached (_add_10).
211017-18:08:08,921 nipype.workflow INFO:
[Job 13] Cached (_add_11).
211017-18:08:10,774 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:10,821 nipype.workflow INFO:
[Job 2] Cached (hello_mapnode.add_1).
211017-18:08:10,868 nipype.workflow INFO:
[Job 14] Cached (_add_10).
211017-18:08:12,778 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:12,825 nipype.workflow INFO:
[Job 15] Cached (_add_11).
211017-18:08:12,839 nipype.workflow INFO:
[Job 16] Cached (_add_10).
211017-18:08:14,779 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:14,812 nipype.workflow INFO:
[Job 4] Cached (hello_mapnode.add_1).
211017-18:08:14,824 nipype.workflow INFO:
[Job 17] Cached (_add_11).
211017-18:08:16,784 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:16,830 nipype.workflow INFO:
[Job 7] Cached (hello_mapnode.add_1).
211017-18:08:16,845 nipype.workflow INFO:
[Job 18] Cached (_add_10).
211017-18:08:18,792 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:18,859 nipype.workflow INFO:
[Job 8] Cached (hello_mapnode.join_scale_data).
211017-18:08:18,874 nipype.workflow INFO:
[Job 19] Cached (_add_11).
211017-18:08:20,792 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:20,825 nipype.workflow INFO:
[Job 10] Cached (hello_mapnode.add_1).
211017-18:08:22,847 nipype.workflow INFO:
[Job 11] Cached (hello_mapnode.join_scale_data).
CPU times: user 516 ms, sys: 52.4 ms, total: 568 ms
Wall time: 22.2 s
wf.base_dir = os.path.join(os.getcwd(), 'alt')
%time eg = wf.run(plugin='MultiProc', plugin_args={'n_procs': 2})
211017-18:08:24,818 nipype.workflow INFO:
Workflow hello_mapnode settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:08:24,864 nipype.workflow INFO:
Running in parallel.
211017-18:08:24,868 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:24,915 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/alt/hello_mapnode/_a_2/a_plus_b".
211017-18:08:24,919 nipype.workflow INFO:
[Node] Setting-up "hello_mapnode.a_plus_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/alt/hello_mapnode/_a_1/a_plus_b".
211017-18:08:24,960 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:24,960 nipype.workflow INFO:
[Node] Running "a_plus_b" ("nipype.interfaces.utility.wrappers.Function")
211017-18:08:25,18 nipype.workflow INFO:
[Node] Finished "hello_mapnode.a_plus_b".
211017-18:08:25,18 nipype.workflow INFO:
[Node] Finished "hello_mapnode.a_plus_b".
211017-18:08:26,871 nipype.workflow INFO:
[Job 0] Completed (hello_mapnode.a_plus_b).
211017-18:08:26,874 nipype.workflow INFO:
[Job 5] Completed (hello_mapnode.a_plus_b).
211017-18:08:26,878 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 2/2.
211017-18:08:26,951 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/alt/hello/_a_2/_b_4/concat_a_b".
211017-18:08:26,951 nipype.workflow INFO:
[Node] Setting-up "hello.concat_a_b" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/alt/hello/_a_2/_b_3/concat_a_b".
211017-18:08:26,995 nipype.workflow INFO:
[Node] Running "concat_a_b" ("nipype.interfaces.utility.wrappers.Function")
%time eg = wf.run(plugin='MultiProc', plugin_args={'n_procs': 2})
Exercise 1¶
Create a workflow to calculate a sum of factorials of numbers from a range between \(n_{min}\) and \(n_{max}\), i.e.:
if \(n_{min}=0\) and \(n_{max}=3\) $\(\sum _{k=0}^{3} k! = 0! + 1! +2! + 3! = 1 + 1 + 2 + 6 = 10\)$
#write your code here
# 1. write 3 functions: one that returns a list of number from a specific range,
# second that returns n! (you can use math.factorial) and third, that sums the elements from a list
# 2. create a workflow and define the working directory
# 3. define 3 nodes using Node and MapNode and connect them within the workflow
# 4. run the workflow and check the results
from nipype import Workflow, Node, MapNode, Function
import os
def range_fun(n_min, n_max):
return list(range(n_min, n_max+1))
def factorial(n):
# print("FACTORIAL, {}".format(n))
import math
return math.factorial(n)
def summing(terms):
return sum(terms)
wf_ex1 = Workflow('ex1')
wf_ex1.base_dir = os.getcwd()
range_nd = Node(Function(input_names=['n_min', 'n_max'],
output_names=['range_list'],
function=range_fun),
name='range_list')
factorial_nd = MapNode(Function(input_names=['n'],
output_names=['fact_out'],
function=factorial),
iterfield=['n'],
name='factorial')
summing_nd = Node(Function(input_names=['terms'],
output_names=['sum_out'],
function=summing),
name='summing')
range_nd.inputs.n_min = 0
range_nd.inputs.n_max = 3
wf_ex1.add_nodes([range_nd])
wf_ex1.connect(range_nd, 'range_list', factorial_nd, 'n')
wf_ex1.connect(factorial_nd, 'fact_out', summing_nd, "terms")
eg = wf_ex1.run()
let’s print all nodes:
eg.nodes()
the final result should be 10:
list(eg.nodes())[2].result.outputs
we can also check the results of two other nodes:
print(list(eg.nodes())[0].result.outputs)
print(list(eg.nodes())[1].result.outputs)
Exercise 2¶
Create a workflow to calculate the following sum for chosen \(n\) and five different values of \(x\): \(0\), \(\frac{1}{2} \pi\), \(\pi\), \(\frac{3}{2} \pi\), and \( 2 \pi\).
\(\sum _{{k=0}}^{{n}}{\frac {(-1)^{k}}{(2k+1)!}}x^{{2k+1}}\quad =x-{\frac {x^{3}}{3!}}+{\frac {x^{5}}{5!}}-\cdots \)
# write your solution here
# 1. write 3 functions: one that returns a list of number from a range between 0 and some n,
# second that returns a term for a specific k, and third, that sums the elements from a list
# 2. create a workflow and define the working directory
# 3. define 3 nodes using Node and MapNode and connect them within the workflow
# 4. use iterables for 4 values of x
# 5. run the workflow and check the final results for every value of x
# we can reuse function from previous exercise, but they need some edits
from nipype import Workflow, Node, MapNode, JoinNode, Function
import os
import math
def range_fun(n_max):
return list(range(n_max+1))
def term(k, x):
import math
fract = math.factorial(2 * k + 1)
polyn = x ** (2 * k + 1)
return (-1)**k * polyn / fract
def summing(terms):
return sum(terms)
wf_ex2 = Workflow('ex2')
wf_ex2.base_dir = os.getcwd()
range_nd = Node(Function(input_names=['n_max'],
output_names=['range_list'],
function=range_fun),
name='range_list')
term_nd = MapNode(Function(input_names=['k', 'x'],
output_names=['term_out'],
function=term),
iterfield=['k'],
name='term')
summing_nd = Node(Function(input_names=['terms'],
output_names=['sum_out'],
function=summing),
name='summing')
range_nd.inputs.n_max = 15
x_list = [0, 0.5 * math.pi, math.pi, 1.5 * math.pi, 2 * math.pi]
term_nd.iterables = ('x', x_list)
wf_ex2.add_nodes([range_nd])
wf_ex2.connect(range_nd, 'range_list', term_nd, 'k')
wf_ex2.connect(term_nd, 'term_out', summing_nd, "terms")
eg = wf_ex2.run()
let’s check all nodes
eg.nodes()
let’s print all results of ex2.summing
print(list(eg.nodes())[2].result.outputs)
print(list(eg.nodes())[4].result.outputs)
print(list(eg.nodes())[6].result.outputs)
print(list(eg.nodes())[8].result.outputs)
print(list(eg.nodes())[10].result.outputs)
Great, we just implemented pretty good Sine function! Those number should be approximately 0, 1, 0, -1 and 0. If they are not, try to increase \(n_max\).
Exercise 2a¶
Use JoinNode to combine results from Exercise 2 in one container, e.g. a dictionary, that takes value \(x\) as a key and the result from summing Node as a value.
# write your code here
# 1. create an additional function that takes 2 lists and combines them into one container, e.g. dictionary
# 2. use JoinNode to define a new node that merges results from Exercise 2 and connect it to the workflow
# 3. run the workflow and check the results of the merging node
def merge_results(results, x):
return dict(zip(x, results))
join_nd = JoinNode(Function(input_names=['results', 'x'],
output_names=['results_cont'],
function=merge_results),
name='merge',
joinsource=term_nd, # this is the node that used iterables for x
joinfield=['results'])
# taking the list of arguments from the previous part
join_nd.inputs.x = x_list
# connecting a new node to the summing_nd
wf_ex2.connect(summing_nd, "sum_out", join_nd, "results")
eg = wf_ex2.run()
let’s print all nodes
eg.nodes()
and results from merge Node:
list(eg.nodes())[1].result.outputs