Nipype Showcase¶
What’s all the hype about Nipype? Is it really that good? Short answer: Yes!
Long answer: … well, let’s consider a very simple fMRI preprocessing workflow that just performs:
slice time correction
motion correction
smoothing
Preparing the preprocessing workflow¶
First, we need to import the main Nipype tools: Node and Workflow
from nipype import Node, Workflow
Now, we can import the interfaces that we want to use for the preprocessing.
from nipype.interfaces.fsl import SliceTimer, MCFLIRT, Smooth
Next, we will put the three interfaces into a node and define the specific input parameters.
# Initiate a node to correct for slice wise acquisition
slicetimer = Node(SliceTimer(index_dir=False,
interleaved=True,
time_repetition=2.5),
name="slicetimer")
# Initiate a node to correct for motion
mcflirt = Node(MCFLIRT(mean_vol=True,
save_plots=True),
name="mcflirt")
# Initiate a node to smooth functional images
smooth = Node(Smooth(fwhm=4), name="smooth")
After creating the nodes, we can now create the preprocessing workflow.
preproc01 = Workflow(name='preproc01', base_dir='.')
Now, we can put all the nodes into this preprocessing workflow. We specify the data flow / execution flow of the workflow by connecting the corresponding nodes to each other.
preproc01.connect([(slicetimer, mcflirt, [('slice_time_corrected_file', 'in_file')]),
(mcflirt, smooth, [('out_file', 'in_file')])])
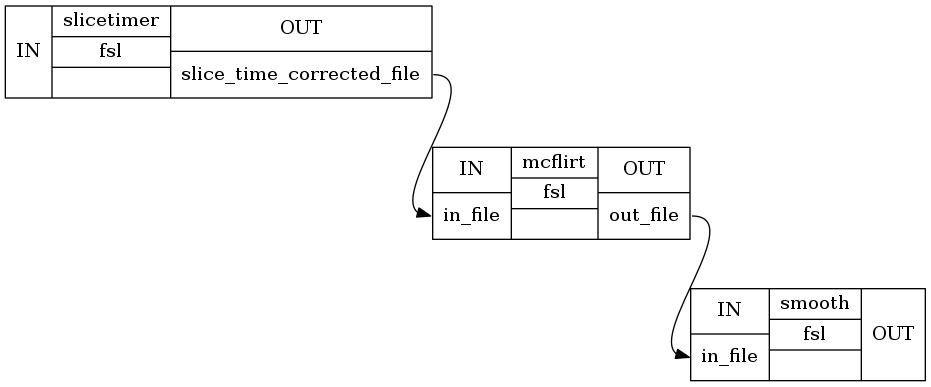
To better understand what we did we can write out the workflow graph and visualize it directly in this notebook.
preproc01.write_graph(graph2use='orig')
211017-18:13:04,666 nipype.workflow INFO:
Generated workflow graph: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/graph.png (graph2use=orig, simple_form=True).
'/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/graph.png'
# Visualize graph
from IPython.display import Image
Image(filename="preproc01/graph_detailed.png")
Run the workflow on one functional image¶
Now, that we’ve created a workflow, let’s run it on a functional image.
For this, we first need to specify the input file of the very first node, i.e. the slicetimer node.
slicetimer.inputs.in_file = '/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz'
To show off Nipype’s parallelization power, let’s run the workflow in parallel, on 5 processors and let’s show the execution time:
%time preproc01.run('MultiProc', plugin_args={'n_procs': 5})
211017-18:13:04,745 nipype.workflow INFO:
Workflow preproc01 settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:13:04,775 nipype.workflow INFO:
Running in parallel.
211017-18:13:04,779 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:13:04,860 nipype.workflow INFO:
[Node] Setting-up "preproc01.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/slicetimer".
211017-18:13:04,894 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:13:06,783 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 4/5.
Currently running:
* preproc01.slicetimer
211017-18:13:18,103 nipype.workflow INFO:
[Node] Finished "preproc01.slicetimer".
211017-18:13:18,792 nipype.workflow INFO:
[Job 0] Completed (preproc01.slicetimer).
211017-18:13:18,796 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:13:18,836 nipype.workflow INFO:
[Node] Setting-up "preproc01.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/mcflirt".
211017-18:13:18,890 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:13:20,796 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 4/5.
Currently running:
* preproc01.mcflirt
211017-18:14:37,994 nipype.workflow INFO:
[Node] Finished "preproc01.mcflirt".
211017-18:14:39,30 nipype.workflow INFO:
[Job 1] Completed (preproc01.mcflirt).
211017-18:14:39,34 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:14:39,76 nipype.workflow INFO:
[Node] Setting-up "preproc01.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/smooth".
211017-18:14:39,131 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 1.699 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:14:41,63 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 4/5.
Currently running:
* preproc01.smooth
211017-18:14:52,247 nipype.workflow INFO:
[Node] Finished "preproc01.smooth".
211017-18:14:53,56 nipype.workflow INFO:
[Job 2] Completed (preproc01.smooth).
211017-18:14:53,59 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
CPU times: user 2.06 s, sys: 139 ms, total: 2.19 s
Wall time: 1min 50s
<networkx.classes.digraph.DiGraph at 0x7f09b197d510>
Conclusion¶
Nice, the whole execution took ~2min. But wait… The parallelization didn’t really help.
That’s true, but because there was no possibility to run the workflow in parallel. Each node depends on the output of the previous node.
Results of preproc01¶
So, what did we get? Let’s look at the output folder preproc01:
!tree preproc01 -I '*js|*json|*pklz|_report|*.dot|*html'
preproc01
├── graph_detailed.png
├── graph.png
├── mcflirt
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
├── slicetimer
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
└── smooth
├── command.txt
└── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
3 directories, 8 files
Rerunning of a workflow¶
Now, for fun. Let’s run the workflow again, but let’s change the fwhm value of the Gaussian smoothing kernel to 2.
smooth.inputs.fwhm = 2
And let’s run the workflow again.
%time preproc01.run('MultiProc', plugin_args={'n_procs': 5})
211017-18:14:55,793 nipype.workflow INFO:
Workflow preproc01 settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:14:55,844 nipype.workflow INFO:
Running in parallel.
211017-18:14:55,848 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:14:55,908 nipype.workflow INFO:
[Job 0] Cached (preproc01.slicetimer).
211017-18:14:57,927 nipype.workflow INFO:
[Job 1] Cached (preproc01.mcflirt).
211017-18:14:59,933 nipype.workflow INFO:
[Node] Outdated cache found for "preproc01.smooth".
211017-18:14:59,978 nipype.workflow INFO:
[Node] Setting-up "preproc01.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/smooth".
211017-18:14:59,989 nipype.workflow INFO:
[Node] Outdated cache found for "preproc01.smooth".
211017-18:15:00,64 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/preproc01/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:15:01,829 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 4/5.
Currently running:
* preproc01.smooth
211017-18:15:10,599 nipype.workflow INFO:
[Node] Finished "preproc01.smooth".
211017-18:15:11,835 nipype.workflow INFO:
[Job 2] Completed (preproc01.smooth).
211017-18:15:11,837 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
CPU times: user 410 ms, sys: 88.2 ms, total: 498 ms
Wall time: 18.1 s
<networkx.classes.digraph.DiGraph at 0x7f09b1928c90>
Conclusion¶
Interesting, now it only took ~15s to execute the whole workflow again. What happened?
As you can see from the log above, Nipype didn’t execute the two nodes slicetimer and mclfirt again. This, because their input values didn’t change from the last execution. The preproc01 workflow therefore only had to rerun the node smooth.
Running a workflow in parallel¶
Ok, ok… Rerunning a workflow again is faster. That’s nice and all, but I want more. You spoke of parallel execution!
We saw that the preproc01 workflow takes about ~2min to execute completely. So, if we would run the workflow on five functional images, it should take about ~10min total. This, of course, assuming the execution will be done sequentially. Now, let’s see how long it takes if we run it in parallel.
# First, let's copy/clone 'preproc01'
preproc02 = preproc01.clone('preproc02')
preproc03 = preproc01.clone('preproc03')
preproc04 = preproc01.clone('preproc04')
preproc05 = preproc01.clone('preproc05')
We now have five different preprocessing workflows. If we want to run them in parallel, we can put them all in another workflow.
metaflow = Workflow(name='metaflow', base_dir='.')
# Now we can add the five preproc workflows to the bigger metaflow
metaflow.add_nodes([preproc01, preproc02, preproc03,
preproc04, preproc05])
Note: We now have a workflow (metaflow), that contains five other workflows (preproc0?), each of them containing three nodes.
To better understand this, let’s visualize this metaflow.
# As before, let's write the graph of the workflow
metaflow.write_graph(graph2use='flat')
211017-18:15:14,299 nipype.workflow INFO:
Generated workflow graph: /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/graph.png (graph2use=flat, simple_form=True).
'/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/graph.png'
# And visualize the graph
from IPython.display import Image
Image(filename="metaflow/graph_detailed.png")
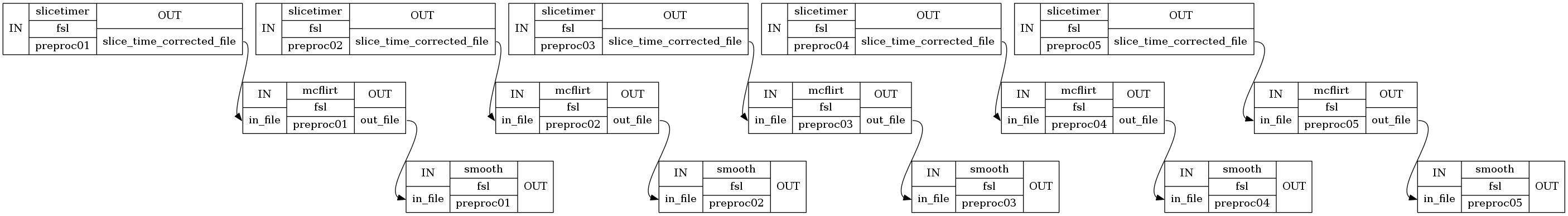
Ah… so now we can see that the metaflow has potential for parallelization. So let’s put it to test
%time metaflow.run('MultiProc', plugin_args={'n_procs': 5})
211017-18:15:14,337 nipype.workflow INFO:
Workflow metaflow settings: ['check', 'execution', 'logging', 'monitoring']
211017-18:15:14,368 nipype.workflow INFO:
Running in parallel.
211017-18:15:14,372 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:15:14,443 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc02.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/slicetimer".
211017-18:15:14,440 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc01.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/slicetimer".
211017-18:15:14,444 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc03.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/slicetimer".
211017-18:15:14,451 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc05.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/slicetimer".
211017-18:15:14,445 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc04.slicetimer" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/slicetimer".
211017-18:15:14,478 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:15:14,480 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:15:14,491 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:15:14,502 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:15:14,523 nipype.workflow INFO:
[Node] Running "slicetimer" ("nipype.interfaces.fsl.preprocess.SliceTimer"), a CommandLine Interface with command:
slicetimer --in=/data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-fingerfootlips_bold.nii.gz --odd --out=/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz --repeat=2.500000
211017-18:15:16,394 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 0.75/1.75, Free processors: 0/5.
Currently running:
* metaflow.preproc05.slicetimer
* metaflow.preproc04.slicetimer
* metaflow.preproc03.slicetimer
* metaflow.preproc02.slicetimer
* metaflow.preproc01.slicetimer
211017-18:15:40,622 nipype.workflow INFO:
[Node] Finished "metaflow.preproc05.slicetimer".
211017-18:15:40,783 nipype.workflow INFO:
[Node] Finished "metaflow.preproc02.slicetimer".
211017-18:15:41,4 nipype.workflow INFO:
[Node] Finished "metaflow.preproc03.slicetimer".
211017-18:15:41,69 nipype.workflow INFO:
[Node] Finished "metaflow.preproc04.slicetimer".
211017-18:15:41,86 nipype.workflow INFO:
[Node] Finished "metaflow.preproc01.slicetimer".
211017-18:15:42,372 nipype.workflow INFO:
[Job 0] Completed (metaflow.preproc01.slicetimer).
211017-18:15:42,376 nipype.workflow INFO:
[Job 3] Completed (metaflow.preproc02.slicetimer).
211017-18:15:42,379 nipype.workflow INFO:
[Job 6] Completed (metaflow.preproc03.slicetimer).
211017-18:15:42,380 nipype.workflow INFO:
[Job 9] Completed (metaflow.preproc04.slicetimer).
211017-18:15:42,384 nipype.workflow INFO:
[Job 12] Completed (metaflow.preproc05.slicetimer).
211017-18:15:42,388 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
211017-18:15:42,437 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc01.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/mcflirt".
211017-18:15:42,438 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc02.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/mcflirt".
211017-18:15:42,440 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc03.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/mcflirt".
211017-18:15:42,442 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc04.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/mcflirt".
211017-18:15:42,452 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc05.mcflirt" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/mcflirt".
211017-18:15:42,515 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:15:42,529 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:15:42,530 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:15:42,531 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:15:42,535 nipype.workflow INFO:
[Node] Running "mcflirt" ("nipype.interfaces.fsl.preprocess.MCFLIRT"), a CommandLine Interface with command:
mcflirt -in /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/slicetimer/sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz -meanvol -out /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -plots
211017-18:15:44,899 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 0.75/1.75, Free processors: 0/5.
Currently running:
* metaflow.preproc05.mcflirt
* metaflow.preproc04.mcflirt
* metaflow.preproc03.mcflirt
* metaflow.preproc02.mcflirt
* metaflow.preproc01.mcflirt
211017-18:18:17,924 nipype.workflow INFO:
[Node] Finished "metaflow.preproc01.mcflirt".
211017-18:18:18,421 nipype.workflow INFO:
[Job 1] Completed (metaflow.preproc01.mcflirt).
211017-18:18:18,430 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 0.95/1.75, Free processors: 1/5.
Currently running:
* metaflow.preproc05.mcflirt
* metaflow.preproc04.mcflirt
* metaflow.preproc03.mcflirt
* metaflow.preproc02.mcflirt
211017-18:18:18,529 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc01.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/smooth".
211017-18:18:18,617 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc01/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:18:20,425 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 0.75/1.75, Free processors: 0/5.
Currently running:
* metaflow.preproc01.smooth
* metaflow.preproc05.mcflirt
* metaflow.preproc04.mcflirt
* metaflow.preproc03.mcflirt
* metaflow.preproc02.mcflirt
211017-18:18:22,605 nipype.workflow INFO:
[Node] Finished "metaflow.preproc05.mcflirt".
211017-18:18:22,940 nipype.workflow INFO:
[Node] Finished "metaflow.preproc03.mcflirt".
211017-18:18:23,5 nipype.workflow INFO:
[Node] Finished "metaflow.preproc02.mcflirt".
211017-18:18:23,686 nipype.workflow INFO:
[Node] Finished "metaflow.preproc04.mcflirt".
211017-18:18:24,426 nipype.workflow INFO:
[Job 4] Completed (metaflow.preproc02.mcflirt).
211017-18:18:24,429 nipype.workflow INFO:
[Job 7] Completed (metaflow.preproc03.mcflirt).
211017-18:18:24,431 nipype.workflow INFO:
[Job 10] Completed (metaflow.preproc04.mcflirt).
211017-18:18:24,436 nipype.workflow INFO:
[Job 13] Completed (metaflow.preproc05.mcflirt).
211017-18:18:24,442 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 4 jobs ready. Free memory (GB): 1.55/1.75, Free processors: 4/5.
Currently running:
* metaflow.preproc01.smooth
211017-18:18:24,498 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc02.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/smooth".
211017-18:18:24,500 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc03.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/smooth".
211017-18:18:24,512 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc05.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/smooth".
211017-18:18:24,506 nipype.workflow INFO:
[Node] Setting-up "metaflow.preproc04.smooth" in "/home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/smooth".
211017-18:18:24,605 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc03/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:18:24,623 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc02/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:18:24,669 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc05/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:18:24,731 nipype.workflow INFO:
[Node] Running "smooth" ("nipype.interfaces.fsl.utils.Smooth"), a CommandLine Interface with command:
fslmaths /home/neuro/workshop_weizmann/workshop/nipype/notebooks/metaflow/preproc04/mcflirt/sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz -kernel gauss 0.849 -fmean sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
211017-18:18:26,434 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 0.75/1.75, Free processors: 0/5.
Currently running:
* metaflow.preproc05.smooth
* metaflow.preproc04.smooth
* metaflow.preproc03.smooth
* metaflow.preproc02.smooth
* metaflow.preproc01.smooth
211017-18:18:38,871 nipype.workflow INFO:
[Node] Finished "metaflow.preproc01.smooth".
211017-18:18:40,516 nipype.workflow INFO:
[Job 2] Completed (metaflow.preproc01.smooth).
211017-18:18:40,522 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 0 jobs ready. Free memory (GB): 0.95/1.75, Free processors: 1/5.
Currently running:
* metaflow.preproc05.smooth
* metaflow.preproc04.smooth
* metaflow.preproc03.smooth
* metaflow.preproc02.smooth
211017-18:18:44,929 nipype.workflow INFO:
[Node] Finished "metaflow.preproc04.smooth".
211017-18:18:45,98 nipype.workflow INFO:
[Node] Finished "metaflow.preproc03.smooth".
211017-18:18:45,350 nipype.workflow INFO:
[Node] Finished "metaflow.preproc02.smooth".
211017-18:18:45,405 nipype.workflow INFO:
[Node] Finished "metaflow.preproc05.smooth".
211017-18:18:46,522 nipype.workflow INFO:
[Job 5] Completed (metaflow.preproc02.smooth).
211017-18:18:46,525 nipype.workflow INFO:
[Job 8] Completed (metaflow.preproc03.smooth).
211017-18:18:46,527 nipype.workflow INFO:
[Job 11] Completed (metaflow.preproc04.smooth).
211017-18:18:46,531 nipype.workflow INFO:
[Job 14] Completed (metaflow.preproc05.smooth).
211017-18:18:46,536 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 1.75/1.75, Free processors: 5/5.
CPU times: user 792 ms, sys: 611 ms, total: 1.4 s
Wall time: 3min 34s
<networkx.classes.digraph.DiGraph at 0x7f09b12c0a90>
This time we can see that Nipype uses all available processors.
And if all went well, the total execution time should still be around ~2min.
That’s why Nipype is so amazing. The days of opening multiple SPMs, FSLs, AFNIs etc. are past!
Results of metaflow¶
!tree metaflow -I '*js|*json|*pklz|_report|*.dot|*html'
metaflow
├── graph_detailed.png
├── graph.png
├── preproc01
│ ├── mcflirt
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
│ ├── slicetimer
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
│ └── smooth
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
├── preproc02
│ ├── mcflirt
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
│ ├── slicetimer
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
│ └── smooth
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
├── preproc03
│ ├── mcflirt
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
│ ├── slicetimer
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
│ └── smooth
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
├── preproc04
│ ├── mcflirt
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
│ ├── slicetimer
│ │ ├── command.txt
│ │ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
│ └── smooth
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
└── preproc05
├── mcflirt
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st_mcf.nii.gz
├── slicetimer
│ ├── command.txt
│ └── sub-01_ses-test_task-fingerfootlips_bold_st.nii.gz
└── smooth
├── command.txt
└── sub-01_ses-test_task-fingerfootlips_bold_st_mcf_smooth.nii.gz
20 directories, 32 files