Hands-on 2: How to create a fMRI analysis workflow¶
The purpose of this section is that you set up a complete fMRI analysis workflow yourself. So that in the end, you are able to perform the analysis from A-Z, i.e. from preprocessing to group analysis. This section will cover the analysis part, the previous section Hands-on 1: Preprocessing handles the preprocessing part.
We will use this opportunity to show you some nice additional interfaces/nodes that might not be relevant to your usual analysis. But it’s always nice to know that they exist. And hopefully, this will encourage you to investigate all other interfaces that Nipype can bring to the tip of your finger.
Important: You will not be able to go through this notebook if you haven’t preprocessed your subjects first.
1st-level Analysis Workflow Structure¶
In this notebook, we will create a workflow that performs 1st-level analysis and normalizes the resulting beta weights to the MNI template. In concrete steps this means:
1. Specify 1st-level model parameters
2. Specify 1st-level contrasts
3. Estimate 1st-level contrasts
4. Normalize 1st-level contrasts
Imports¶
It’s always best to have all relevant module imports at the beginning of your script. So let’s import what we most certainly need.
from nilearn import plotting
%matplotlib inline
# Get the Node and Workflow object
from nipype import Node, Workflow
# Specify which SPM to use
from nipype.interfaces.matlab import MatlabCommand
MatlabCommand.set_default_paths('/opt/spm12-r7219/spm12_mcr/spm12')
Note: Ideally you would also put the imports of all the interfaces that you use here at the top. But as we will develop the workflow step by step, we can also import the relevant modules as we go.
Create Nodes and Workflow connections¶
Let’s create all the nodes that we need! Make sure to specify all relevant inputs and keep in mind which ones you later on need to connect in your pipeline.
Workflow for the 1st-level analysis¶
We recommend to create the workflow and establish all its connections at a later place in your script. This helps to have everything nicely together. But for this hands-on example, it makes sense to establish the connections between the nodes as we go.
And for this, we first need to create a workflow:
# Create the workflow here
# Hint: use 'base_dir' to specify where to store the working directory
analysis1st = Workflow(name='work_1st', base_dir='/output/')
Specify 1st-level model parameters (stimuli onsets, duration, etc.)¶
The specify the 1st-level model we need the subject-specific onset times and duration of the stimuli. Luckily, as we are working with a BIDS dataset, this information is nicely stored in a tsv file:
import pandas as pd
trialinfo = pd.read_table('/home/neuro/workshop/data/ds000114/task-fingerfootlips_events.tsv')
trialinfo
| onset | duration | weight | trial_type | |
|---|---|---|---|---|
| 0 | 10 | 15.0 | 1 | Finger |
| 1 | 40 | 15.0 | 1 | Foot |
| 2 | 70 | 15.0 | 1 | Lips |
| 3 | 100 | 15.0 | 1 | Finger |
| 4 | 130 | 15.0 | 1 | Foot |
| 5 | 160 | 15.0 | 1 | Lips |
| 6 | 190 | 15.0 | 1 | Finger |
| 7 | 220 | 15.0 | 1 | Foot |
| 8 | 250 | 15.0 | 1 | Lips |
| 9 | 280 | 15.0 | 1 | Finger |
| 10 | 310 | 15.0 | 1 | Foot |
| 11 | 340 | 15.0 | 1 | Lips |
| 12 | 370 | 15.0 | 1 | Finger |
| 13 | 400 | 15.0 | 1 | Foot |
| 14 | 430 | 15.0 | 1 | Lips |
Using pandas is probably the quickest and easiest ways to aggregate stimuli information per condition.
for group in trialinfo.groupby('trial_type'):
print(group)
print("")
('Finger', onset duration weight trial_type
0 10 15.0 1 Finger
3 100 15.0 1 Finger
6 190 15.0 1 Finger
9 280 15.0 1 Finger
12 370 15.0 1 Finger)
('Foot', onset duration weight trial_type
1 40 15.0 1 Foot
4 130 15.0 1 Foot
7 220 15.0 1 Foot
10 310 15.0 1 Foot
13 400 15.0 1 Foot)
('Lips', onset duration weight trial_type
2 70 15.0 1 Lips
5 160 15.0 1 Lips
8 250 15.0 1 Lips
11 340 15.0 1 Lips
14 430 15.0 1 Lips)
To create a GLM model, Nipype needs an list of Bunch objects per session. As we only have one session, our object needs to look as follows:
[Bunch(conditions=['Finger', 'Foot', 'Lips'],
durations=[[15.0, 15.0, 15.0, 15.0, 15.0],
[15.0, 15.0, 15.0, 15.0, 15.0],
[15.0, 15.0, 15.0, 15.0, 15.0]],
onsets=[[10, 100, 190, 280, 370],
[40, 130, 220, 310, 400],
[70, 160, 250, 340, 430]]
)]
For more information see either the official documnetation or the nipype_tutorial example.
So, let’s create this Bunch object that we then can use for the GLM model.
import pandas as pd
from nipype.interfaces.base import Bunch
trialinfo = pd.read_table('/home/neuro/workshop/data/ds000114/task-fingerfootlips_events.tsv')
conditions = []
onsets = []
durations = []
for group in trialinfo.groupby('trial_type'):
conditions.append(group[0])
onsets.append(list(group[1].onset -10)) # subtracting 10s due to removing of 4 dummy scans
durations.append(group[1].duration.tolist())
subject_info = [Bunch(conditions=conditions,
onsets=onsets,
durations=durations,
)]
subject_info
[Bunch(conditions=['Finger', 'Foot', 'Lips'],
durations=[[15.0, 15.0, 15.0, 15.0, 15.0],
[15.0, 15.0, 15.0, 15.0, 15.0],
[15.0, 15.0, 15.0, 15.0, 15.0]],
onsets=[[0, 90, 180, 270, 360],
[30, 120, 210, 300, 390],
[60, 150, 240, 330, 420]])]
Good! Now we can create the node that will create the SPM model. For this we will be using SpecifySPMModel. As a reminder the TR of the acquisition is 2.5s and we want to use a high pass filter of 128.
from nipype.algorithms.modelgen import SpecifySPMModel
# Initiate the SpecifySPMModel node here
modelspec = Node(SpecifySPMModel(concatenate_runs=False,
input_units='secs',
output_units='secs',
time_repetition=2.5,
high_pass_filter_cutoff=128,
subject_info=subject_info),
name="modelspec")
This node will also need some additional inputs, such as the preprocessed functional images, the motion parameters etc. We will specify those once we take care of the workflow data input stream.
Specify 1st-level contrasts¶
To do any GLM analysis, we need to also define the contrasts that we want to investigate. If we recap, we had three different conditions in the fingerfootlips task in this dataset:
finger
foot
lips
Therefore, we could create the following contrasts (seven T-contrasts and two F-contrasts):
# Condition names
condition_names = ['Finger', 'Foot', 'Lips']
# Contrasts
cont01 = ['average', 'T', condition_names, [1/3., 1/3., 1/3.]]
cont02 = ['Finger', 'T', condition_names, [1, 0, 0]]
cont03 = ['Foot', 'T', condition_names, [0, 1, 0]]
cont04 = ['Lips', 'T', condition_names, [0, 0, 1]]
cont05 = ['Finger < others','T', condition_names, [-1, 0.5, 0.5]]
cont06 = ['Foot < others', 'T', condition_names, [0.5, -1, 0.5]]
cont07 = ['Lips > others', 'T', condition_names, [-0.5, -0.5, 1]]
cont08 = ['activation', 'F', [cont02, cont03, cont04]]
cont09 = ['differences', 'F', [cont05, cont06, cont07]]
contrast_list = [cont01, cont02, cont03, cont04, cont05, cont06, cont07, cont08, cont09]
Estimate 1st-level contrasts¶
Before we can estimate the 1st-level contrasts, we first need to create the 1st-level design. Here you can also specify what kind of basis function you want (HRF, FIR, Fourier, etc.), if you want to use time and dispersion derivatives and how you want to model the serial correlation.
In this example, I propose that you use an HRF basis function, that we model time derivatives and that we model the serial correlation with AR(1).
from nipype.interfaces.spm import Level1Design
# Initiate the Level1Design node here
level1design = Node(Level1Design(bases={'hrf': {'derivs': [1, 0]}},
timing_units='secs',
interscan_interval=2.5,
model_serial_correlations='AR(1)'),
name="level1design")
Now that we have the Model Specification and 1st-Level Design node, we can connect them to each other:
# Connect the two nodes here
analysis1st.connect([(modelspec, level1design, [('session_info',
'session_info')])])
Now we need to estimate the model. I recommend that you’ll use a Classical: 1 method to estimate the model.
from nipype.interfaces.spm import EstimateModel
# Initiate the EstimateModel node here
level1estimate = Node(EstimateModel(estimation_method={'Classical': 1}),
name="level1estimate")
Now we can connect the 1st-Level Design node with the model estimation node.
# Connect the two nodes here
analysis1st.connect([(level1design, level1estimate, [('spm_mat_file',
'spm_mat_file')])])
Now that we estimate the model, we can estimate the contrasts. Don’t forget to feed the list of contrast we specify above to this node.
from nipype.interfaces.spm import EstimateContrast
# Initiate the EstimateContrast node here
level1conest = Node(EstimateContrast(contrasts=contrast_list),
name="level1conest")
Now we can connect the model estimation node with the contrast estimation node.
# Connect the two nodes here
analysis1st.connect([(level1estimate, level1conest, [('spm_mat_file',
'spm_mat_file'),
('beta_images',
'beta_images'),
('residual_image',
'residual_image')])])
Normalize 1st-level contrasts¶
Now that the contrasts were estimated in subject space we can put them into a common reference space by normalizing them to a specific template. In this case, we will be using SPM12’s Normalize routine and normalize to the SPM12 tissue probability map TPM.nii.
At this step, you can also specify the voxel resolution of the output volumes. If you don’t specify it, it will normalize to a voxel resolution of 2x2x2mm. As a training exercise, set the voxel resolution to 4x4x4mm.
from nipype.interfaces.spm import Normalize12
# Location of the template
template = '/opt/spm12-r7219/spm12_mcr/spm12/tpm/TPM.nii'
# Initiate the Normalize12 node here
normalize = Node(Normalize12(jobtype='estwrite',
tpm=template,
write_voxel_sizes=[4, 4, 4]
),
name="normalize")
Now we can connect the estimated contrasts to normalization node.
# Connect the nodes here
analysis1st.connect([(level1conest, normalize, [('con_images',
'apply_to_files')])
])
Datainput with SelectFiles and iterables¶
As in the preprocessing hands-on, we will again be using SelectFiles and iterables. So, what do we need?
From the preprocessing pipeline, we need the functional images, the motion parameters and the list of outliers. Also, for the normalization, we need the subject-specific anatomy.
# Import the SelectFiles
from nipype import SelectFiles
# String template with {}-based strings
templates = {'anat': '/home/neuro/workshop/data/ds000114/sub-{subj_id}/ses-test/anat/sub-{subj_id}_ses-test_T1w.nii.gz',
'func': '/output/datasink_handson/preproc/sub-{subj_id}_detrend.nii.gz',
'mc_param': '/output/datasink_handson/preproc/sub-{subj_id}.par',
'outliers': '/output/datasink_handson/preproc/art.sub-{subj_id}_outliers.txt'
}
# Create SelectFiles node
sf = Node(SelectFiles(templates, sort_filelist=True),
name='selectfiles')
Now we can specify over which subjects the workflow should iterate. As we preprocessed only subjects ‘02’, ‘03’, ‘04’, ‘07’, ‘08’ and ‘09’ , we can only them for this analysis.
# list of subject identifiers
subject_list = ['02', '03', '04', '07', '08', '09']
sf.iterables = [('subj_id', subject_list)]
Gunzip Node¶
SPM12 can accept NIfTI files as input, but online if they are not compressed (‘unzipped’). Therefore, we need to use a Gunzip node to unzip the detrend file and another one to unzip the anatomy image, before we can feed it to the model specification node.
from nipype.algorithms.misc import Gunzip
# Initiate the two Gunzip node here
gunzip_anat = Node(Gunzip(), name='gunzip_anat')
gunzip_func = Node(Gunzip(), name='gunzip_func')
And as a final step, we just need to connect this SelectFiles node to the rest of the workflow.
# Connect SelectFiles node to the other nodes here
analysis1st.connect([(sf, gunzip_anat, [('anat', 'in_file')]),
(sf, gunzip_func, [('func', 'in_file')]),
(gunzip_anat, normalize, [('out_file', 'image_to_align')]),
(gunzip_func, modelspec, [('out_file', 'functional_runs')]),
(sf, modelspec, [('mc_param', 'realignment_parameters'),
('outliers', 'outlier_files'),
])
])
Data output with DataSink¶
Now, before we run the workflow, let’s again specify a Datasink folder to only keep those files that we want to keep.
from nipype.interfaces.io import DataSink
# Initiate DataSink node here
# Initiate the datasink node
output_folder = 'datasink_handson'
datasink = Node(DataSink(base_directory='/output/',
container=output_folder),
name="datasink")
## Use the following substitutions for the DataSink output
substitutions = [('_subj_id_', 'sub-')]
datasink.inputs.substitutions = substitutions
Now the next step is to specify all the output that we want to keep in our output folder output. Probably best to keep are the:
SPM.mat file and the spmT and spmF files from the contrast estimation node
normalized betas and anatomy
# Connect nodes to datasink here
analysis1st.connect([(level1conest, datasink, [('spm_mat_file', '1stLevel.@spm_mat'),
('spmT_images', '1stLevel.@T'),
('spmF_images', '1stLevel.@F'),
]),
(normalize, datasink, [('normalized_files', 'normalized.@files'),
('normalized_image', 'normalized.@image'),
]),
])
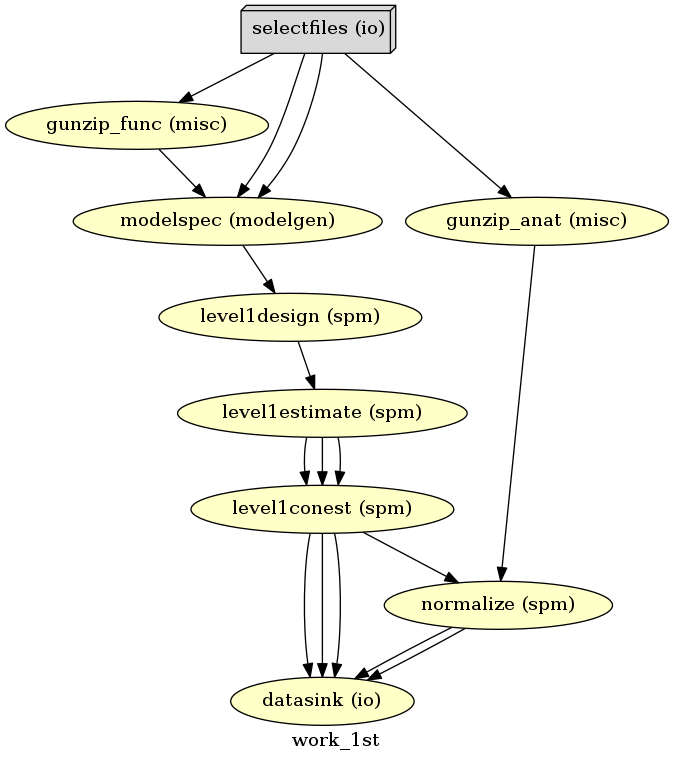
Visualize the workflow¶
Now that the workflow is finished, let’s visualize it again.
# Create 1st-level analysis output graph
analysis1st.write_graph(graph2use='colored', format='png', simple_form=True)
# Visualize the graph
from IPython.display import Image
Image(filename='/output/work_1st/graph.png')
211018-10:40:01,932 nipype.workflow INFO:
Generated workflow graph: /output/work_1st/graph.png (graph2use=colored, simple_form=True).
Run the Workflow¶
Now that everything is ready, we can run the 1st-level analysis workflow. Change n_procs to the number of jobs/cores you want to use.
analysis1st.run('MultiProc', plugin_args={'n_procs': 4})
211018-10:40:01,977 nipype.workflow INFO:
Workflow work_1st settings: ['check', 'execution', 'logging', 'monitoring']
211018-10:40:02,42 nipype.workflow INFO:
Running in parallel.
211018-10:40:02,48 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 6 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:02,191 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_08/selectfiles".
211018-10:40:02,196 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_04/selectfiles".
211018-10:40:02,195 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_07/selectfiles".
211018-10:40:02,189 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_09/selectfiles".
211018-10:40:02,210 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:02,220 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:02,226 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:02,229 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:02,237 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:02,241 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:02,248 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:02,249 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:04,57 nipype.workflow INFO:
[Job 0] Completed (work_1st.selectfiles).
211018-10:40:04,60 nipype.workflow INFO:
[Job 9] Completed (work_1st.selectfiles).
211018-10:40:04,63 nipype.workflow INFO:
[Job 18] Completed (work_1st.selectfiles).
211018-10:40:04,66 nipype.workflow INFO:
[Job 27] Completed (work_1st.selectfiles).
211018-10:40:04,69 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 10 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:04,232 nipype.workflow INFO:
[Job 1] Cached (work_1st.gunzip_func).
211018-10:40:04,237 nipype.workflow INFO:
[Job 6] Cached (work_1st.gunzip_anat).
211018-10:40:04,244 nipype.workflow INFO:
[Job 10] Cached (work_1st.gunzip_func).
211018-10:40:04,248 nipype.workflow INFO:
[Job 15] Cached (work_1st.gunzip_anat).
211018-10:40:06,59 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 8 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:06,128 nipype.workflow INFO:
[Job 2] Cached (work_1st.modelspec).
211018-10:40:06,136 nipype.workflow INFO:
[Job 11] Cached (work_1st.modelspec).
211018-10:40:06,141 nipype.workflow INFO:
[Job 19] Cached (work_1st.gunzip_func).
211018-10:40:06,146 nipype.workflow INFO:
[Job 24] Cached (work_1st.gunzip_anat).
211018-10:40:08,67 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:08,151 nipype.workflow INFO:
[Job 3] Cached (work_1st.level1design).
211018-10:40:08,173 nipype.workflow INFO:
[Job 12] Cached (work_1st.level1design).
211018-10:40:08,182 nipype.workflow INFO:
[Job 20] Cached (work_1st.modelspec).
211018-10:40:08,185 nipype.workflow INFO:
[Job 28] Cached (work_1st.gunzip_func).
211018-10:40:10,132 nipype.workflow INFO:
[Job 4] Cached (work_1st.level1estimate).
211018-10:40:10,137 nipype.workflow INFO:
[Job 13] Cached (work_1st.level1estimate).
211018-10:40:10,163 nipype.workflow INFO:
[Job 21] Cached (work_1st.level1design).
211018-10:40:10,172 nipype.workflow INFO:
[Job 29] Cached (work_1st.modelspec).
211018-10:40:12,146 nipype.workflow INFO:
[Job 5] Cached (work_1st.level1conest).
211018-10:40:12,161 nipype.workflow INFO:
[Job 14] Cached (work_1st.level1conest).
211018-10:40:12,167 nipype.workflow INFO:
[Job 22] Cached (work_1st.level1estimate).
211018-10:40:12,191 nipype.workflow INFO:
[Job 30] Cached (work_1st.level1design).
211018-10:40:14,146 nipype.workflow INFO:
[Job 7] Cached (work_1st.normalize).
211018-10:40:14,158 nipype.workflow INFO:
[Job 16] Cached (work_1st.normalize).
211018-10:40:14,170 nipype.workflow INFO:
[Job 23] Cached (work_1st.level1conest).
211018-10:40:14,176 nipype.workflow INFO:
[Job 31] Cached (work_1st.level1estimate).
211018-10:40:16,151 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_09/datasink".
211018-10:40:16,159 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:16,163 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/SPM.mat -> /output/datasink_handson/1stLevel/sub-09/SPM.mat
211018-10:40:16,166 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0001.nii
211018-10:40:16,169 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0002.nii
211018-10:40:16,167 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_08/datasink".
211018-10:40:16,174 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0003.nii
211018-10:40:16,176 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0004.nii
211018-10:40:16,180 nipype.workflow INFO:
[Job 25] Cached (work_1st.normalize).
211018-10:40:16,182 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:16,185 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0005.nii
211018-10:40:16,189 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/SPM.mat -> /output/datasink_handson/1stLevel/sub-08/SPM.mat
211018-10:40:16,191 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0006.nii
211018-10:40:16,194 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0001.nii
211018-10:40:16,194 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-09/spmT_0007.nii
211018-10:40:16,197 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0002.nii
211018-10:40:16,200 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-09/spmF_0008.nii
211018-10:40:16,200 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0003.nii
211018-10:40:16,203 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-09/spmF_0009.nii
211018-10:40:16,205 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0004.nii
211018-10:40:16,210 nipype.workflow INFO:
[Job 32] Cached (work_1st.level1conest).
211018-10:40:16,208 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-09/spmF_0008.nii
211018-10:40:16,209 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0005.nii
211018-10:40:16,211 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0006.nii
211018-10:40:16,212 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_09/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-09/spmF_0009.nii
211018-10:40:16,214 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0001.nii -> /output/datasink_handson/normalized/sub-09/wcon_0001.nii
211018-10:40:16,213 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-08/spmT_0007.nii
211018-10:40:16,216 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0002.nii -> /output/datasink_handson/normalized/sub-09/wcon_0002.nii
211018-10:40:16,219 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0003.nii -> /output/datasink_handson/normalized/sub-09/wcon_0003.nii
211018-10:40:16,218 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-08/spmF_0008.nii
211018-10:40:16,221 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0004.nii -> /output/datasink_handson/normalized/sub-09/wcon_0004.nii
211018-10:40:16,221 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-08/spmF_0009.nii
211018-10:40:16,224 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0005.nii -> /output/datasink_handson/normalized/sub-09/wcon_0005.nii
211018-10:40:16,224 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-08/spmF_0008.nii
211018-10:40:16,225 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0006.nii -> /output/datasink_handson/normalized/sub-09/wcon_0006.nii
211018-10:40:16,225 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_08/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-08/spmF_0009.nii
211018-10:40:16,227 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0001.nii -> /output/datasink_handson/normalized/sub-08/wcon_0001.nii
211018-10:40:16,227 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wcon_0007.nii -> /output/datasink_handson/normalized/sub-09/wcon_0007.nii
211018-10:40:16,229 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0002.nii -> /output/datasink_handson/normalized/sub-08/wcon_0002.nii
211018-10:40:16,229 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wess_0008.nii -> /output/datasink_handson/normalized/sub-09/wess_0008.nii
211018-10:40:16,231 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0003.nii -> /output/datasink_handson/normalized/sub-08/wcon_0003.nii
211018-10:40:16,231 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wess_0009.nii -> /output/datasink_handson/normalized/sub-09/wess_0009.nii
211018-10:40:16,233 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0004.nii -> /output/datasink_handson/normalized/sub-08/wcon_0004.nii
211018-10:40:16,235 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_09/wsub-09_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-09/wsub-09_ses-test_T1w.nii
211018-10:40:16,235 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0005.nii -> /output/datasink_handson/normalized/sub-08/wcon_0005.nii
211018-10:40:16,237 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0006.nii -> /output/datasink_handson/normalized/sub-08/wcon_0006.nii
211018-10:40:16,242 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:16,245 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wcon_0007.nii -> /output/datasink_handson/normalized/sub-08/wcon_0007.nii
211018-10:40:16,246 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wess_0008.nii -> /output/datasink_handson/normalized/sub-08/wess_0008.nii
211018-10:40:16,248 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wess_0009.nii -> /output/datasink_handson/normalized/sub-08/wess_0009.nii
211018-10:40:16,250 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_08/wsub-08_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-08/wsub-08_ses-test_T1w.nii
211018-10:40:16,256 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:18,82 nipype.workflow INFO:
[Job 8] Completed (work_1st.datasink).
211018-10:40:18,87 nipype.workflow INFO:
[Job 17] Completed (work_1st.datasink).
211018-10:40:18,97 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:18,254 nipype.workflow INFO:
[Job 33] Cached (work_1st.gunzip_anat).
211018-10:40:18,253 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_07/datasink".
211018-10:40:18,260 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:18,262 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/SPM.mat -> /output/datasink_handson/1stLevel/sub-07/SPM.mat
211018-10:40:18,266 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0001.nii
211018-10:40:18,268 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_03/selectfiles".
211018-10:40:18,272 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0002.nii
211018-10:40:18,274 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0003.nii
211018-10:40:18,278 nipype.workflow INFO:
[Node] Setting-up "work_1st.selectfiles" in "/output/work_1st/_subj_id_02/selectfiles".
211018-10:40:18,276 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0004.nii
211018-10:40:18,275 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:18,286 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:18,289 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0005.nii
211018-10:40:18,293 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0006.nii
211018-10:40:18,298 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-07/spmT_0007.nii
211018-10:40:18,301 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:18,301 nipype.workflow INFO:
[Node] Finished "work_1st.selectfiles".
211018-10:40:18,304 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-07/spmF_0008.nii
211018-10:40:18,308 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-07/spmF_0009.nii
211018-10:40:18,311 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-07/spmF_0008.nii
211018-10:40:18,313 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_07/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-07/spmF_0009.nii
211018-10:40:18,317 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0001.nii -> /output/datasink_handson/normalized/sub-07/wcon_0001.nii
211018-10:40:18,321 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0002.nii -> /output/datasink_handson/normalized/sub-07/wcon_0002.nii
211018-10:40:18,326 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0003.nii -> /output/datasink_handson/normalized/sub-07/wcon_0003.nii
211018-10:40:18,330 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0004.nii -> /output/datasink_handson/normalized/sub-07/wcon_0004.nii
211018-10:40:18,335 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0005.nii -> /output/datasink_handson/normalized/sub-07/wcon_0005.nii
211018-10:40:18,339 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0006.nii -> /output/datasink_handson/normalized/sub-07/wcon_0006.nii
211018-10:40:18,342 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wcon_0007.nii -> /output/datasink_handson/normalized/sub-07/wcon_0007.nii
211018-10:40:18,346 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wess_0008.nii -> /output/datasink_handson/normalized/sub-07/wess_0008.nii
211018-10:40:18,349 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wess_0009.nii -> /output/datasink_handson/normalized/sub-07/wess_0009.nii
211018-10:40:18,353 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_07/wsub-07_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-07/wsub-07_ses-test_T1w.nii
211018-10:40:18,365 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:20,84 nipype.workflow INFO:
[Job 26] Completed (work_1st.datasink).
211018-10:40:20,86 nipype.workflow INFO:
[Job 36] Completed (work_1st.selectfiles).
211018-10:40:20,88 nipype.workflow INFO:
[Job 45] Completed (work_1st.selectfiles).
211018-10:40:20,93 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:20,184 nipype.workflow INFO:
[Job 34] Cached (work_1st.normalize).
211018-10:40:20,192 nipype.workflow INFO:
[Job 37] Cached (work_1st.gunzip_func).
211018-10:40:20,196 nipype.workflow INFO:
[Job 42] Cached (work_1st.gunzip_anat).
211018-10:40:20,201 nipype.workflow INFO:
[Job 46] Cached (work_1st.gunzip_func).
211018-10:40:22,91 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:22,173 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_04/datasink".
211018-10:40:22,183 nipype.workflow INFO:
[Job 38] Cached (work_1st.modelspec).
211018-10:40:22,183 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:22,187 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/SPM.mat -> /output/datasink_handson/1stLevel/sub-04/SPM.mat
211018-10:40:22,190 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0001.nii
211018-10:40:22,194 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0002.nii
211018-10:40:22,197 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0003.nii
211018-10:40:22,200 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0004.nii
211018-10:40:22,201 nipype.workflow INFO:
[Job 47] Cached (work_1st.modelspec).
211018-10:40:22,203 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0005.nii
211018-10:40:22,207 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0006.nii
211018-10:40:22,209 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-04/spmT_0007.nii
211018-10:40:22,213 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-04/spmF_0008.nii
211018-10:40:22,216 nipype.workflow INFO:
[Job 51] Cached (work_1st.gunzip_anat).
211018-10:40:22,215 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-04/spmF_0009.nii
211018-10:40:22,219 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-04/spmF_0008.nii
211018-10:40:22,221 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_04/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-04/spmF_0009.nii
211018-10:40:22,224 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0001.nii -> /output/datasink_handson/normalized/sub-04/wcon_0001.nii
211018-10:40:22,227 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0002.nii -> /output/datasink_handson/normalized/sub-04/wcon_0002.nii
211018-10:40:22,230 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0003.nii -> /output/datasink_handson/normalized/sub-04/wcon_0003.nii
211018-10:40:22,234 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0004.nii -> /output/datasink_handson/normalized/sub-04/wcon_0004.nii
211018-10:40:22,236 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0005.nii -> /output/datasink_handson/normalized/sub-04/wcon_0005.nii
211018-10:40:22,239 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0006.nii -> /output/datasink_handson/normalized/sub-04/wcon_0006.nii
211018-10:40:22,241 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wcon_0007.nii -> /output/datasink_handson/normalized/sub-04/wcon_0007.nii
211018-10:40:22,244 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wess_0008.nii -> /output/datasink_handson/normalized/sub-04/wess_0008.nii
211018-10:40:22,247 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wess_0009.nii -> /output/datasink_handson/normalized/sub-04/wess_0009.nii
211018-10:40:22,250 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_04/wsub-04_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-04/wsub-04_ses-test_T1w.nii
211018-10:40:22,257 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:24,90 nipype.workflow INFO:
[Job 35] Completed (work_1st.datasink).
211018-10:40:24,93 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:24,196 nipype.workflow INFO:
[Job 39] Cached (work_1st.level1design).
211018-10:40:24,221 nipype.workflow INFO:
[Job 48] Cached (work_1st.level1design).
211018-10:40:26,172 nipype.workflow INFO:
[Job 40] Cached (work_1st.level1estimate).
211018-10:40:26,182 nipype.workflow INFO:
[Job 49] Cached (work_1st.level1estimate).
211018-10:40:28,176 nipype.workflow INFO:
[Job 41] Cached (work_1st.level1conest).
211018-10:40:28,193 nipype.workflow INFO:
[Job 50] Cached (work_1st.level1conest).
211018-10:40:30,194 nipype.workflow INFO:
[Job 43] Cached (work_1st.normalize).
211018-10:40:30,211 nipype.workflow INFO:
[Job 52] Cached (work_1st.normalize).
211018-10:40:32,188 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_03/datasink".
211018-10:40:32,199 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:32,202 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/SPM.mat -> /output/datasink_handson/1stLevel/sub-03/SPM.mat
211018-10:40:32,206 nipype.workflow INFO:
[Node] Setting-up "work_1st.datasink" in "/output/work_1st/_subj_id_02/datasink".
211018-10:40:32,207 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0001.nii
211018-10:40:32,212 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0002.nii
211018-10:40:32,216 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0003.nii
211018-10:40:32,218 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0004.nii
211018-10:40:32,221 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0005.nii
211018-10:40:32,220 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:40:32,227 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0006.nii
211018-10:40:32,227 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/SPM.mat -> /output/datasink_handson/1stLevel/sub-02/SPM.mat
211018-10:40:32,230 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-03/spmT_0007.nii
211018-10:40:32,230 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0001.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0001.nii
211018-10:40:32,234 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-03/spmF_0008.nii
211018-10:40:32,233 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0002.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0002.nii
211018-10:40:32,238 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0003.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0003.nii
211018-10:40:32,241 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-03/spmF_0009.nii
211018-10:40:32,242 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0004.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0004.nii
211018-10:40:32,244 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-03/spmF_0008.nii
211018-10:40:32,246 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0005.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0005.nii
211018-10:40:32,248 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_03/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-03/spmF_0009.nii
211018-10:40:32,248 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0006.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0006.nii
211018-10:40:32,251 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmT_0007.nii -> /output/datasink_handson/1stLevel/sub-02/spmT_0007.nii
211018-10:40:32,251 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0001.nii -> /output/datasink_handson/normalized/sub-03/wcon_0001.nii
211018-10:40:32,254 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-02/spmF_0008.nii
211018-10:40:32,257 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0002.nii -> /output/datasink_handson/normalized/sub-03/wcon_0002.nii
211018-10:40:32,257 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-02/spmF_0009.nii
211018-10:40:32,260 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmF_0008.nii -> /output/datasink_handson/1stLevel/sub-02/spmF_0008.nii
211018-10:40:32,263 nipype.interface INFO:
sub: /output/datasink_handson/1stLevel/_subj_id_02/spmF_0009.nii -> /output/datasink_handson/1stLevel/sub-02/spmF_0009.nii
211018-10:40:32,260 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0003.nii -> /output/datasink_handson/normalized/sub-03/wcon_0003.nii
211018-10:40:32,267 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0004.nii -> /output/datasink_handson/normalized/sub-03/wcon_0004.nii
211018-10:40:32,266 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0001.nii -> /output/datasink_handson/normalized/sub-02/wcon_0001.nii
211018-10:40:32,270 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0005.nii -> /output/datasink_handson/normalized/sub-03/wcon_0005.nii
211018-10:40:32,271 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0002.nii -> /output/datasink_handson/normalized/sub-02/wcon_0002.nii
211018-10:40:32,273 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0006.nii -> /output/datasink_handson/normalized/sub-03/wcon_0006.nii
211018-10:40:32,274 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0003.nii -> /output/datasink_handson/normalized/sub-02/wcon_0003.nii
211018-10:40:32,277 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wcon_0007.nii -> /output/datasink_handson/normalized/sub-03/wcon_0007.nii
211018-10:40:32,278 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0004.nii -> /output/datasink_handson/normalized/sub-02/wcon_0004.nii
211018-10:40:32,281 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0005.nii -> /output/datasink_handson/normalized/sub-02/wcon_0005.nii
211018-10:40:32,283 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wess_0008.nii -> /output/datasink_handson/normalized/sub-03/wess_0008.nii
211018-10:40:32,284 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0006.nii -> /output/datasink_handson/normalized/sub-02/wcon_0006.nii
211018-10:40:32,286 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wess_0009.nii -> /output/datasink_handson/normalized/sub-03/wess_0009.nii
211018-10:40:32,289 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_03/wsub-03_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-03/wsub-03_ses-test_T1w.nii
211018-10:40:32,289 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wcon_0007.nii -> /output/datasink_handson/normalized/sub-02/wcon_0007.nii
211018-10:40:32,292 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wess_0008.nii -> /output/datasink_handson/normalized/sub-02/wess_0008.nii
211018-10:40:32,294 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wess_0009.nii -> /output/datasink_handson/normalized/sub-02/wess_0009.nii
211018-10:40:32,298 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:32,298 nipype.interface INFO:
sub: /output/datasink_handson/normalized/_subj_id_02/wsub-02_ses-test_T1w.nii -> /output/datasink_handson/normalized/sub-02/wsub-02_ses-test_T1w.nii
211018-10:40:32,304 nipype.workflow INFO:
[Node] Finished "work_1st.datasink".
211018-10:40:34,115 nipype.workflow INFO:
[Job 44] Completed (work_1st.datasink).
211018-10:40:34,117 nipype.workflow INFO:
[Job 53] Completed (work_1st.datasink).
211018-10:40:34,120 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
<networkx.classes.digraph.DiGraph at 0x7fdc41c338d0>
Visualize results¶
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
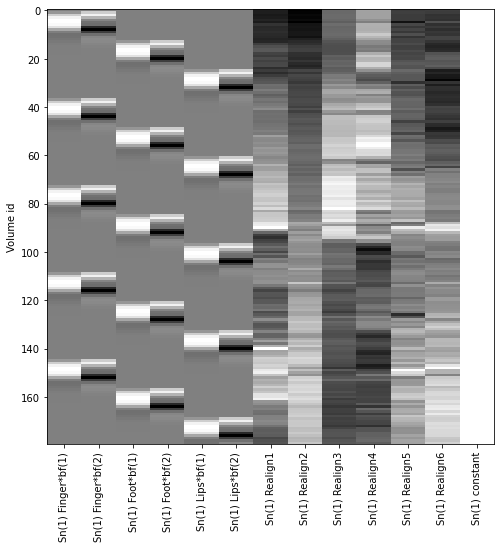
First, let’s look at the 1st-level Design Matrix of subject one, to verify that everything is as it should be.
from scipy.io import loadmat
# Using scipy's loadmat function we can access SPM.mat
spmmat = loadmat('/output/datasink_handson/1stLevel/sub-07/SPM.mat',
struct_as_record=False)
The design matrix and the names of the regressors are a bit hidden in the spmmat variable, but they can be accessed as follows:
designMatrix = spmmat['SPM'][0][0].xX[0][0].X
names = [i[0] for i in spmmat['SPM'][0][0].xX[0][0].name[0]]
Now before we can plot it, we just need to normalize the desing matrix in such a way, that each column has a maximum amplitude of 1. This is just for visualization purposes, otherwise the rotation parameters with their rather small values will not show up in the figure.
normed_design = designMatrix / np.abs(designMatrix).max(axis=0)
And we’re ready to plot the design matrix.
fig, ax = plt.subplots(figsize=(8, 8))
plt.imshow(normed_design, aspect='auto', cmap='gray', interpolation='none')
ax.set_ylabel('Volume id')
ax.set_xticks(np.arange(len(names)))
ax.set_xticklabels(names, rotation=90);
Now that we’re happy with the design matrix, let’s look how well the normalization worked.
import nibabel as nb
from nilearn.plotting import plot_anat
from nilearn.plotting import plot_glass_brain
# Load GM probability map of TPM.nii
img = nb.load('/opt/spm12-r7219/spm12_mcr/spm12/tpm/TPM.nii')
GM_template = nb.Nifti1Image(img.get_fdata()[..., 0], img.affine, img.header)
# Plot normalized subject anatomy
display = plot_anat('/output/datasink_handson/normalized/sub-07/wsub-07_ses-test_T1w.nii',
dim=-0.1)
# Overlay in edges GM map
display.add_edges(GM_template)
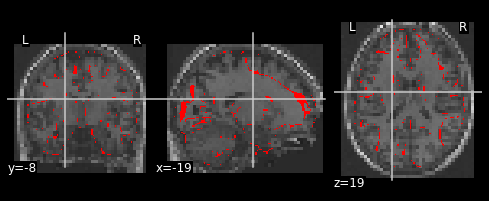
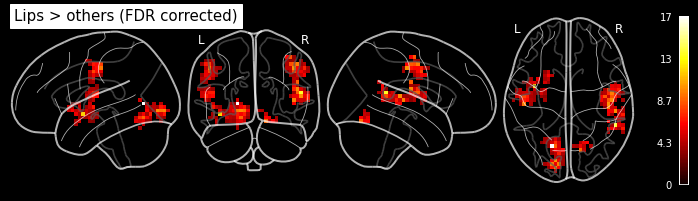
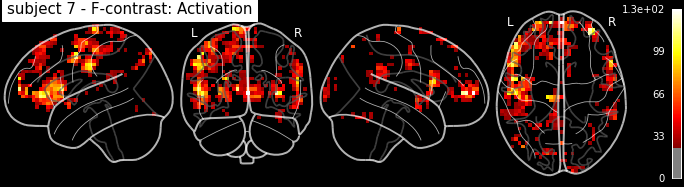
Let’s look at the contrasts of one subject that we’ve just computed. In particular the F-contrast.
plot_glass_brain('/output/datasink_handson/normalized/sub-07/wess_0008.nii',
output_file="/output/datasink_handson/normalized/sub-07/f-contr_activation.png",
colorbar=True, display_mode='lyrz', black_bg=True, threshold=25,
title='subject 7 - F-contrast: Activation');
Image(filename='/output/datasink_handson/normalized/sub-07/f-contr_activation.png')
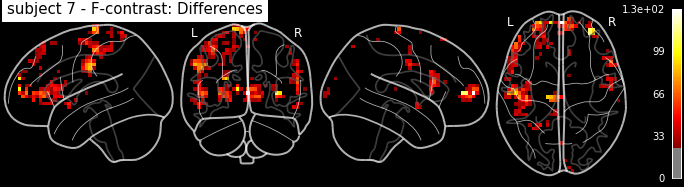
plot_glass_brain('/output/datasink_handson/normalized/sub-07/wess_0009.nii',
output_file="/output/datasink_handson/normalized/sub-07/f-contr_differences.png",
colorbar=True, display_mode='lyrz', black_bg=True, threshold=25,
title='subject 7 - F-contrast: Differences');
Image(filename='/output/datasink_handson/normalized/sub-07/f-contr_differences.png')
2nd-level Analysis Workflow Structure¶
Last but not least, the group level analysis. This example will also directly include thresholding of the output, as well as some visualization.
Imports¶
To make sure that the necessary imports are done, here they are again:
# Get the Node and Workflow object
from nipype import Node, Workflow
# Specify which SPM to use
from nipype.interfaces.matlab import MatlabCommand
MatlabCommand.set_default_paths('/opt/spm12-r7219/spm12_mcr/spm12')
Create Nodes and Workflow connections¶
Now we should know this part very well.
Workflow for the 2nd-level analysis¶
# Create the workflow here
# Hint: use 'base_dir' to specify where to store the working directory
analysis2nd = Workflow(name='work_2nd', base_dir='/output/')
2nd-Level Design¶
This step depends on your study design and the tests you want to perform. If you’re using SPM to do the group analysis, you have the liberty to choose between a factorial design, a multiple regression design, one-sample T-Test design, a paired T-Test design or a two-sample T-Test design.
For the current example, we will be using a one sample T-Test design.
from nipype.interfaces.spm import OneSampleTTestDesign
# Initiate the OneSampleTTestDesign node here
onesamplettestdes = Node(OneSampleTTestDesign(), name="onesampttestdes")
The next two steps are the same as for the 1st-level design, i.e. estimation of the model followed by estimation of the contrasts.
from nipype.interfaces.spm import EstimateModel, EstimateContrast
# Initiate the EstimateModel and the EstimateContrast node here
level2estimate = Node(EstimateModel(estimation_method={'Classical': 1}),
name="level2estimate")
level2conestimate = Node(EstimateContrast(group_contrast=True),
name="level2conestimate")
To finish the EstimateContrast node, we also need to specify which contrast should be computed. For a 2nd-level one sample t-test design, this is rather straightforward:
cont01 = ['Group', 'T', ['mean'], [1]]
level2conestimate.inputs.contrasts = [cont01]
Now, let’s connect those three design nodes to each other.
# Connect OneSampleTTestDesign, EstimateModel and EstimateContrast here
analysis2nd.connect([(onesamplettestdes, level2estimate, [('spm_mat_file',
'spm_mat_file')]),
(level2estimate, level2conestimate, [('spm_mat_file',
'spm_mat_file'),
('beta_images',
'beta_images'),
('residual_image',
'residual_image')])
])
Thresholding of output contrast¶
And to close, we will use SPM Threshold. With this routine, we can set a specific voxel threshold (i.e. p<0.001) and apply an FDR cluster threshold (i.e. p<0.05).
As we only have 5 subjects, I recommend to set the voxel threshold to 0.01 and to leave the cluster threshold at 0.05.
from nipype.interfaces.spm import Threshold
level2thresh = Node(Threshold(contrast_index=1,
use_topo_fdr=True,
use_fwe_correction=False,
extent_threshold=0,
height_threshold=0.01,
height_threshold_type='p-value',
extent_fdr_p_threshold=0.05),
name="level2thresh")
# Connect the Threshold node to the EstimateContrast node here
analysis2nd.connect([(level2conestimate, level2thresh, [('spm_mat_file',
'spm_mat_file'),
('spmT_images',
'stat_image'),
])
])
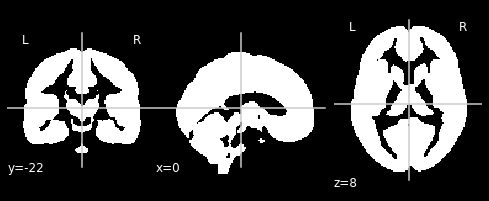
Gray Matter Mask¶
We could run our 2nd-level workflow as it is. All the major nodes are there. But I nonetheless suggest that we use a gray matter mask to restrict the analysis to only gray matter voxels.
In the 1st-level analysis, we normalized to SPM12’s TPM.nii tissue probability atlas. Therefore, we could just take the gray matter probability map of this TPM.nii image (the first volume) and threshold it at a certain probability value to get a binary mask. This can of course also all be done in Nipype, but sometimes the direct bash code is quicker:
%%bash
TEMPLATE='/opt/spm12-r7219/spm12_mcr/spm12/tpm/TPM.nii'
# Extract the first volume with `fslroi`
fslroi $TEMPLATE GM_PM.nii.gz 0 1
# Threshold the probability mask at 10%
fslmaths GM_PM.nii -thr 0.10 -bin /output/datasink_handson/GM_mask.nii.gz
# Unzip the mask and delete the GM_PM.nii file
gunzip /output/datasink_handson/GM_mask.nii.gz
rm GM_PM.nii.gz
gzip: /output/datasink_handson/GM_mask.nii already exists; not overwritten
Let’s take a look at this mask:
from nilearn.plotting import plot_anat
%matplotlib inline
plot_anat('/output/datasink_handson/GM_mask.nii', dim=-1)
<nilearn.plotting.displays.OrthoSlicer at 0x7fdc32609b10>
Now we just need to specify this binary mask as an explicit_mask_file for the one sample T-test node.
onesamplettestdes.inputs.explicit_mask_file = '/output/datasink_handson/GM_mask.nii'
Datainput with SelectFiles and iterables¶
We will again be using SelectFiles and iterables.
So, what do we need? Actually, just the 1st-level contrasts of all subjects, separated by contrast number.
# Import the SelectFiles
from nipype import SelectFiles
# String template with {}-based strings
templates = {'cons': '/output/datasink_handson/normalized/sub-*/w*_{cont_id}.nii'}
# Create SelectFiles node
sf = Node(SelectFiles(templates, sort_filelist=True),
name='selectfiles')
We are using * to tell SelectFiles that it can grab all available subjects and any contrast, with a specific contrast id, independnet if it’s an t-contrast (con) or an F-contrast (ess) contrast.
So, let’s specify over which contrast the workflow should iterate.
# list of contrast identifiers
contrast_id_list = ['0001', '0002', '0003', '0004', '0005', '0006', '0007']
sf.iterables = [('cont_id', contrast_id_list)]
Now we need to connect the SelectFiles to the OneSampleTTestDesign node.
analysis2nd.connect([(sf, onesamplettestdes, [('cons', 'in_files')])])
Data output with DataSink¶
Now, before we run the workflow, let’s again specify a Datasink folder to only keep those files that we want to keep.
from nipype.interfaces.io import DataSink
# Initiate DataSink node here
# Initiate the datasink node
output_folder = 'datasink_handson'
datasink = Node(DataSink(base_directory='/output/',
container=output_folder),
name="datasink")
## Use the following substitutions for the DataSink output
substitutions = [('_cont_id_', 'con_')]
datasink.inputs.substitutions = substitutions
Now the next step is to specify all the output that we want to keep in our output folder output. Probably best to keep are the:
the SPM.mat file and the spmT images from the
EstimateContrastnodethe thresholded spmT images from the
Thresholdnode
# Connect nodes to datasink here
analysis2nd.connect([(level2conestimate, datasink, [('spm_mat_file',
'2ndLevel.@spm_mat'),
('spmT_images',
'2ndLevel.@T'),
('con_images',
'2ndLevel.@con')]),
(level2thresh, datasink, [('thresholded_map',
'2ndLevel.@threshold')])
])
Visualize the workflow¶
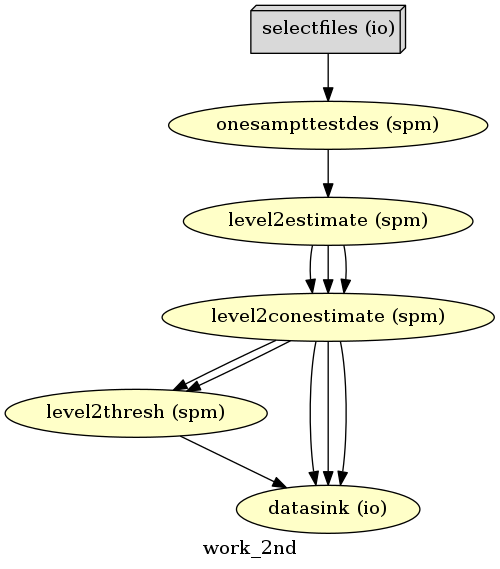
And we’re good to go. Let’s first take a look at the workflow.
# Create 1st-level analysis output graph
analysis2nd.write_graph(graph2use='colored', format='png', simple_form=True)
# Visualize the graph
from IPython.display import Image
Image(filename='/output/work_2nd/graph.png')
211018-10:40:41,575 nipype.workflow INFO:
Generated workflow graph: /output/work_2nd/graph.png (graph2use=colored, simple_form=True).
Run the Workflow¶
Now that everything is ready, we can run the 2nd-level analysis workflow. Change n_procs to the number of jobs/cores you want to use.
analysis2nd.run('MultiProc', plugin_args={'n_procs': 4})
211018-10:40:41,611 nipype.workflow INFO:
Workflow work_2nd settings: ['check', 'execution', 'logging', 'monitoring']
211018-10:40:41,656 nipype.workflow INFO:
Running in parallel.
211018-10:40:41,660 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:41,786 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0005/selectfiles".
211018-10:40:41,786 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0004/selectfiles".
211018-10:40:41,786 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0006/selectfiles".
211018-10:40:41,786 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0007/selectfiles".
211018-10:40:41,797 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:41,797 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:41,796 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:41,802 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:40:41,810 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:40:41,811 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:40:41,812 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:40:41,813 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:40:43,665 nipype.workflow INFO:
[Job 0] Completed (work_2nd.selectfiles).
211018-10:40:43,669 nipype.workflow INFO:
[Job 6] Completed (work_2nd.selectfiles).
211018-10:40:43,671 nipype.workflow INFO:
[Job 12] Completed (work_2nd.selectfiles).
211018-10:40:43,673 nipype.workflow INFO:
[Job 18] Completed (work_2nd.selectfiles).
211018-10:40:43,676 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:43,822 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0007/onesampttestdes".
211018-10:40:43,822 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0006/onesampttestdes".
211018-10:40:43,824 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0004/onesampttestdes".
211018-10:40:43,824 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0005/onesampttestdes".
211018-10:40:43,837 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:40:43,838 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:40:43,840 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:40:43,844 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:40:45,666 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 3 jobs ready. Free memory (GB): 4.43/5.23, Free processors: 0/4.
Currently running:
* work_2nd.onesampttestdes
* work_2nd.onesampttestdes
* work_2nd.onesampttestdes
* work_2nd.onesampttestdes
211018-10:40:53,808 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:40:53,816 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:40:53,827 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:40:53,827 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:40:55,683 nipype.workflow INFO:
[Job 1] Completed (work_2nd.onesampttestdes).
211018-10:40:55,686 nipype.workflow INFO:
[Job 7] Completed (work_2nd.onesampttestdes).
211018-10:40:55,688 nipype.workflow INFO:
[Job 13] Completed (work_2nd.onesampttestdes).
211018-10:40:55,690 nipype.workflow INFO:
[Job 19] Completed (work_2nd.onesampttestdes).
211018-10:40:55,693 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:40:55,775 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0007/level2estimate".
211018-10:40:55,776 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0006/level2estimate".
211018-10:40:55,778 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0004/level2estimate".
211018-10:40:55,777 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0005/level2estimate".
211018-10:40:55,791 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:40:55,790 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:40:55,792 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:40:55,792 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:40:57,686 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 3 jobs ready. Free memory (GB): 4.43/5.23, Free processors: 0/4.
Currently running:
* work_2nd.level2estimate
* work_2nd.level2estimate
* work_2nd.level2estimate
* work_2nd.level2estimate
211018-10:41:08,249 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:08,450 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:08,506 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:08,589 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:09,701 nipype.workflow INFO:
[Job 2] Completed (work_2nd.level2estimate).
211018-10:41:09,703 nipype.workflow INFO:
[Job 8] Completed (work_2nd.level2estimate).
211018-10:41:09,705 nipype.workflow INFO:
[Job 14] Completed (work_2nd.level2estimate).
211018-10:41:09,707 nipype.workflow INFO:
[Job 20] Completed (work_2nd.level2estimate).
211018-10:41:09,710 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:09,841 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0007/level2conestimate".
211018-10:41:09,843 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0006/level2conestimate".
211018-10:41:09,845 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0005/level2conestimate".
211018-10:41:09,848 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0004/level2conestimate".
211018-10:41:09,863 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:09,864 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:09,870 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:09,866 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:11,705 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 3 jobs ready. Free memory (GB): 4.43/5.23, Free processors: 0/4.
Currently running:
* work_2nd.level2conestimate
* work_2nd.level2conestimate
* work_2nd.level2conestimate
* work_2nd.level2conestimate
211018-10:41:18,534 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:41:18,669 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:41:18,674 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:41:18,687 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:41:19,714 nipype.workflow INFO:
[Job 3] Completed (work_2nd.level2conestimate).
211018-10:41:19,717 nipype.workflow INFO:
[Job 9] Completed (work_2nd.level2conestimate).
211018-10:41:19,720 nipype.workflow INFO:
[Job 15] Completed (work_2nd.level2conestimate).
211018-10:41:19,723 nipype.workflow INFO:
[Job 21] Completed (work_2nd.level2conestimate).
211018-10:41:19,728 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:19,807 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0007/level2thresh".
211018-10:41:19,807 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0006/level2thresh".
211018-10:41:19,809 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0005/level2thresh".
211018-10:41:19,809 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0004/level2thresh".
211018-10:41:19,823 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:41:19,823 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:41:19,823 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:41:19,824 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:41:21,715 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 3 jobs ready. Free memory (GB): 4.43/5.23, Free processors: 0/4.
Currently running:
* work_2nd.level2thresh
* work_2nd.level2thresh
* work_2nd.level2thresh
* work_2nd.level2thresh
211018-10:41:22,579 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:41:22,579 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:41:22,633 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:41:22,664 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:41:23,718 nipype.workflow INFO:
[Job 4] Completed (work_2nd.level2thresh).
211018-10:41:23,722 nipype.workflow INFO:
[Job 10] Completed (work_2nd.level2thresh).
211018-10:41:23,724 nipype.workflow INFO:
[Job 16] Completed (work_2nd.level2thresh).
211018-10:41:23,728 nipype.workflow INFO:
[Job 22] Completed (work_2nd.level2thresh).
211018-10:41:23,731 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 7 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:23,819 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0006/datasink".
211018-10:41:23,816 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0007/datasink".
211018-10:41:23,819 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0005/datasink".
211018-10:41:23,819 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0004/datasink".
211018-10:41:23,835 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:41:23,835 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:41:23,838 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0004/SPM.mat -> /output/datasink_handson/2ndLevel/con_0004/SPM.mat
211018-10:41:23,836 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:41:23,841 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0007/SPM.mat -> /output/datasink_handson/2ndLevel/con_0007/SPM.mat
211018-10:41:23,844 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0006/SPM.mat -> /output/datasink_handson/2ndLevel/con_0006/SPM.mat
211018-10:41:23,837 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:41:23,848 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0007/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0007/spmT_0001.nii
211018-10:41:23,851 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0006/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0006/spmT_0001.nii
211018-10:41:23,853 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0007/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0007/con_0001.nii
211018-10:41:23,855 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0004/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0004/spmT_0001.nii
211018-10:41:23,854 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0006/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0006/con_0001.nii
211018-10:41:23,855 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0005/SPM.mat -> /output/datasink_handson/2ndLevel/con_0005/SPM.mat
211018-10:41:23,857 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0007/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0007/spmT_0001_thr.nii
211018-10:41:23,859 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0006/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0006/spmT_0001_thr.nii
211018-10:41:23,862 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0005/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0005/spmT_0001.nii
211018-10:41:23,861 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0004/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0004/con_0001.nii
211018-10:41:23,866 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0004/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0004/spmT_0001_thr.nii
211018-10:41:23,870 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:41:23,870 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:41:23,868 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0005/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0005/con_0001.nii
211018-10:41:23,875 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0005/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0005/spmT_0001_thr.nii
211018-10:41:23,878 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:41:23,885 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:41:25,727 nipype.workflow INFO:
[Job 5] Completed (work_2nd.datasink).
211018-10:41:25,732 nipype.workflow INFO:
[Job 11] Completed (work_2nd.datasink).
211018-10:41:25,739 nipype.workflow INFO:
[Job 17] Completed (work_2nd.datasink).
211018-10:41:25,742 nipype.workflow INFO:
[Job 23] Completed (work_2nd.datasink).
211018-10:41:25,749 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:25,864 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0003/selectfiles".
211018-10:41:25,864 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0001/selectfiles".
211018-10:41:25,864 nipype.workflow INFO:
[Node] Setting-up "work_2nd.selectfiles" in "/output/work_2nd/_cont_id_0002/selectfiles".
211018-10:41:25,868 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:41:25,869 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:41:25,871 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
211018-10:41:25,886 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:41:25,885 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:41:25,889 nipype.workflow INFO:
[Node] Finished "work_2nd.selectfiles".
211018-10:41:27,728 nipype.workflow INFO:
[Job 24] Completed (work_2nd.selectfiles).
211018-10:41:27,734 nipype.workflow INFO:
[Job 30] Completed (work_2nd.selectfiles).
211018-10:41:27,736 nipype.workflow INFO:
[Job 36] Completed (work_2nd.selectfiles).
211018-10:41:27,740 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:27,837 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0003/onesampttestdes".
211018-10:41:27,838 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0002/onesampttestdes".
211018-10:41:27,838 nipype.workflow INFO:
[Node] Setting-up "work_2nd.onesampttestdes" in "/output/work_2nd/_cont_id_0001/onesampttestdes".
211018-10:41:27,850 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:41:27,850 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:41:27,851 nipype.workflow INFO:
[Node] Running "onesampttestdes" ("nipype.interfaces.spm.model.OneSampleTTestDesign")
211018-10:41:29,729 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 4.63/5.23, Free processors: 1/4.
Currently running:
* work_2nd.onesampttestdes
* work_2nd.onesampttestdes
* work_2nd.onesampttestdes
211018-10:41:33,887 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:41:33,887 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:41:33,929 nipype.workflow INFO:
[Node] Finished "work_2nd.onesampttestdes".
211018-10:41:35,734 nipype.workflow INFO:
[Job 25] Completed (work_2nd.onesampttestdes).
211018-10:41:35,736 nipype.workflow INFO:
[Job 31] Completed (work_2nd.onesampttestdes).
211018-10:41:35,737 nipype.workflow INFO:
[Job 37] Completed (work_2nd.onesampttestdes).
211018-10:41:35,741 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:35,830 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0003/level2estimate".
211018-10:41:35,831 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0002/level2estimate".
211018-10:41:35,831 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2estimate" in "/output/work_2nd/_cont_id_0001/level2estimate".
211018-10:41:35,845 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:41:35,845 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:41:35,846 nipype.workflow INFO:
[Node] Running "level2estimate" ("nipype.interfaces.spm.model.EstimateModel")
211018-10:41:37,738 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 4.63/5.23, Free processors: 1/4.
Currently running:
* work_2nd.level2estimate
* work_2nd.level2estimate
* work_2nd.level2estimate
211018-10:41:45,141 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:45,309 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:45,316 nipype.workflow INFO:
[Node] Finished "work_2nd.level2estimate".
211018-10:41:45,747 nipype.workflow INFO:
[Job 26] Completed (work_2nd.level2estimate).
211018-10:41:45,749 nipype.workflow INFO:
[Job 32] Completed (work_2nd.level2estimate).
211018-10:41:45,752 nipype.workflow INFO:
[Job 38] Completed (work_2nd.level2estimate).
211018-10:41:45,756 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:41:45,872 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0003/level2conestimate".
211018-10:41:45,872 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0002/level2conestimate".
211018-10:41:45,872 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2conestimate" in "/output/work_2nd/_cont_id_0001/level2conestimate".
211018-10:41:45,895 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:45,895 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:45,895 nipype.workflow INFO:
[Node] Running "level2conestimate" ("nipype.interfaces.spm.model.EstimateContrast")
211018-10:41:47,749 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 4.63/5.23, Free processors: 1/4.
Currently running:
* work_2nd.level2conestimate
* work_2nd.level2conestimate
* work_2nd.level2conestimate
211018-10:42:03,337 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:42:03,337 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:42:03,382 nipype.workflow INFO:
[Node] Finished "work_2nd.level2conestimate".
211018-10:42:03,848 nipype.workflow INFO:
[Job 27] Completed (work_2nd.level2conestimate).
211018-10:42:03,849 nipype.workflow INFO:
[Job 33] Completed (work_2nd.level2conestimate).
211018-10:42:03,851 nipype.workflow INFO:
[Job 39] Completed (work_2nd.level2conestimate).
211018-10:42:03,855 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:42:03,956 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0001/level2thresh".
211018-10:42:03,955 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0002/level2thresh".
211018-10:42:03,956 nipype.workflow INFO:
[Node] Setting-up "work_2nd.level2thresh" in "/output/work_2nd/_cont_id_0003/level2thresh".
211018-10:42:03,969 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:42:03,969 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:42:03,969 nipype.workflow INFO:
[Node] Running "level2thresh" ("nipype.interfaces.spm.model.Threshold")
211018-10:42:05,863 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 4.63/5.23, Free processors: 1/4.
Currently running:
* work_2nd.level2thresh
* work_2nd.level2thresh
* work_2nd.level2thresh
211018-10:42:07,190 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:42:07,260 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:42:07,294 nipype.workflow INFO:
[Node] Finished "work_2nd.level2thresh".
211018-10:42:07,866 nipype.workflow INFO:
[Job 28] Completed (work_2nd.level2thresh).
211018-10:42:07,868 nipype.workflow INFO:
[Job 34] Completed (work_2nd.level2thresh).
211018-10:42:07,870 nipype.workflow INFO:
[Job 40] Completed (work_2nd.level2thresh).
211018-10:42:07,873 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
211018-10:42:07,964 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0002/datasink".
211018-10:42:07,964 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0003/datasink".
211018-10:42:07,967 nipype.workflow INFO:
[Node] Setting-up "work_2nd.datasink" in "/output/work_2nd/_cont_id_0001/datasink".
211018-10:42:07,979 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:42:07,980 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:42:07,980 nipype.workflow INFO:
[Node] Running "datasink" ("nipype.interfaces.io.DataSink")
211018-10:42:07,983 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0003/SPM.mat -> /output/datasink_handson/2ndLevel/con_0003/SPM.mat
211018-10:42:07,983 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0001/SPM.mat -> /output/datasink_handson/2ndLevel/con_0001/SPM.mat
211018-10:42:07,987 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0002/SPM.mat -> /output/datasink_handson/2ndLevel/con_0002/SPM.mat
211018-10:42:07,993 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0003/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0003/spmT_0001.nii
211018-10:42:07,996 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0001/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0001/spmT_0001.nii
211018-10:42:07,999 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0003/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0003/con_0001.nii
211018-10:42:07,998 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0002/spmT_0001.nii -> /output/datasink_handson/2ndLevel/con_0002/spmT_0001.nii
211018-10:42:08,7 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0003/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0003/spmT_0001_thr.nii
211018-10:42:08,12 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0001/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0001/con_0001.nii
211018-10:42:08,15 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:42:08,15 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0002/con_0001.nii -> /output/datasink_handson/2ndLevel/con_0002/con_0001.nii
211018-10:42:08,28 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0002/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0002/spmT_0001_thr.nii
211018-10:42:08,22 nipype.interface INFO:
sub: /output/datasink_handson/2ndLevel/_cont_id_0001/spmT_0001_thr.nii -> /output/datasink_handson/2ndLevel/con_0001/spmT_0001_thr.nii
211018-10:42:08,35 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:42:08,37 nipype.workflow INFO:
[Node] Finished "work_2nd.datasink".
211018-10:42:09,872 nipype.workflow INFO:
[Job 29] Completed (work_2nd.datasink).
211018-10:42:09,875 nipype.workflow INFO:
[Job 35] Completed (work_2nd.datasink).
211018-10:42:09,879 nipype.workflow INFO:
[Job 41] Completed (work_2nd.datasink).
211018-10:42:09,882 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 5.23/5.23, Free processors: 4/4.
<networkx.classes.digraph.DiGraph at 0x7fdc491ad410>
Visualize results¶
Let’s take a look at the results. Keep in mind that we only have N=6 subjects and that we set the voxel threshold to a very liberal p<0.01. Interpretation of the results should, therefore, be taken with a lot of caution.
from nilearn.plotting import plot_glass_brain
%matplotlib inline
out_path = '/output/datasink_handson/2ndLevel/'
plot_glass_brain(out_path + 'con_0001/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='average (FDR corrected)');
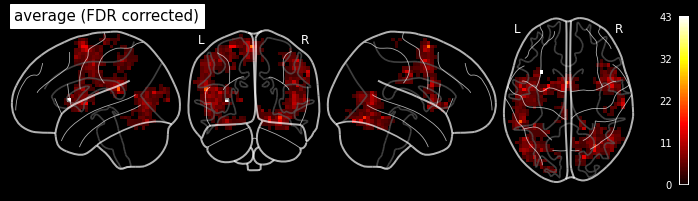
plot_glass_brain(out_path + 'con_0002/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Finger (FDR corrected)');
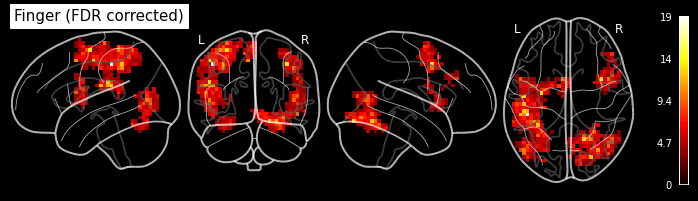
plot_glass_brain(out_path + 'con_0003/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Foot (FDR corrected)');
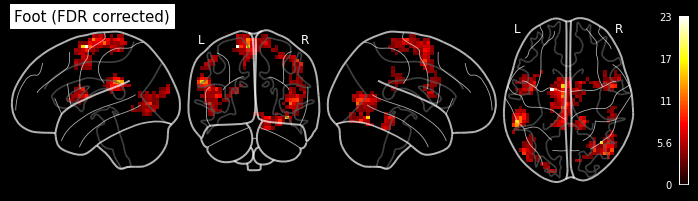
plot_glass_brain(out_path + 'con_0004/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Lips (FDR corrected)');
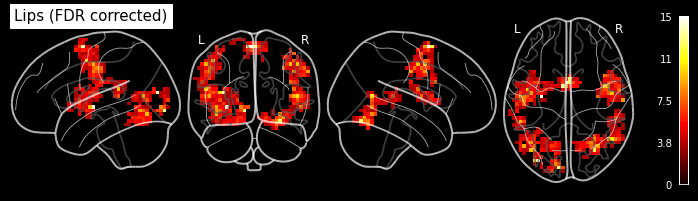
plot_glass_brain(out_path + 'con_0005/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Finger < others (FDR corrected)');
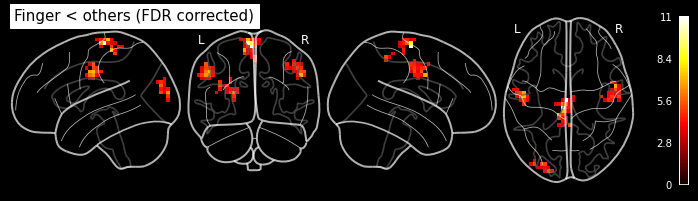
plot_glass_brain(out_path + 'con_0006/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Foot < others (FDR corrected)');
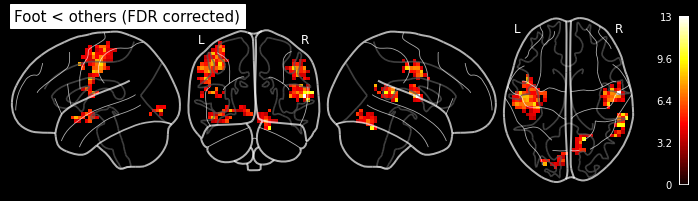
plot_glass_brain(out_path + 'con_0007/spmT_0001_thr.nii', display_mode='lyrz',
black_bg=True, colorbar=True, title='Lips > others (FDR corrected)');